5 - BioMed Central
advertisement
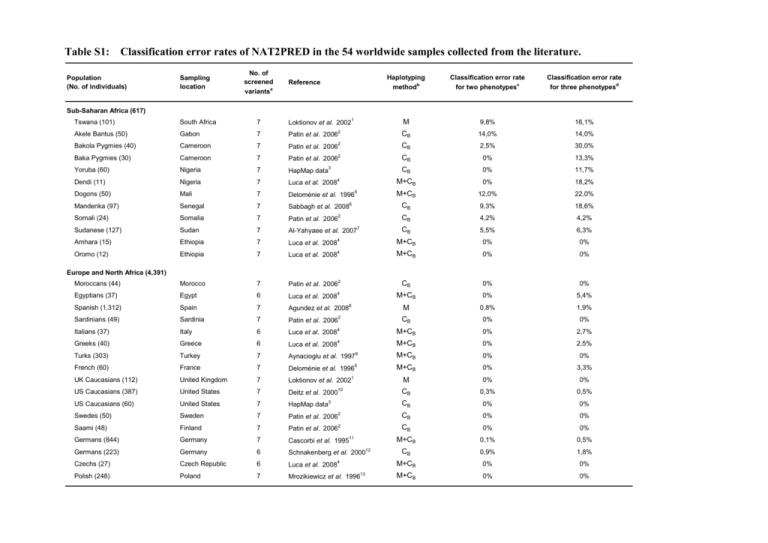
Table S1: Classification error rates of NAT2PRED in the 54 worldwide samples collected from the literature. Population (No. of individuals) Sampling location No. of screened Reference a variants Haplotyping Classification error rate Classification error rate methodb for two phenotypesc for three phenotypesd Sub-Saharan Africa (617) Tswana (101) South Africa 7 Loktionov et al. 20021 M 9,8% 16,1% Akele Bantus (50) Gabon 7 Patin et al. 20062 CB 14,0% 14,0% Bakola Pygmies (40) Cameroon 7 Patin et al. 20062 CB 2,5% 30,0% Baka Pygmies (30) Cameroon 7 2 CB 0% 13,3% Patin et al. 2006 3 Yoruba (60) Nigeria 7 HapMap data Dendi (11) Nigeria 7 Luca et al. 20084 Dogons (50) Mali 7 5 Deloménie et al. 1996 6 CB 0% 11,7% M+CB 0% 18,2% M+CB 12,0% 22,0% 18,6% Mandenka (97) Senegal 7 Sabbagh et al. 2008 CB 9,3% Somali (24) Somalia 7 Patin et al. 20062 CB 4,2% 4,2% Sudanese (127) Sudan 7 Al-Yahyaee et al. 20077 CB 5,5% 6,3% Amhara (15) Ethiopia 7 Luca et al. 20084 M+CB 0% 0% Oromo (12) Ethiopia 7 Luca et al. 20084 M+CB 0% 0% Moroccans (44) Morocco 7 Patin et al. 20062 CB 0% 0% Egyptians (37) Egypt 6 Luca et al. 20084 M+CB 0% 5,4% Spanish (1,312) Spain 7 Agundez et al. 20088 M 0,8% 1,9% Sardinians (49) Sardinia 7 Patin et al. 20062 CB 0% 0% Italians (37) Italy 6 Luca et al. 20084 M+CB 0% 2,7% Greeks (40) Greece 6 Luca et al. 20084 2,5% Europe and North Africa (4,391) M+CB 0% 9 Turks (303) Turkey 7 Aynacioglu et al. 1997 M+CB 0% 0% French (60) France 7 Deloménie et al. 19965 M+CB 0% 3,3% UK Caucasians (112) United Kingdom 7 Loktionov et al. 20021 M 0% 0% 10 US Caucasians (387) United States 7 Deitz et al. 2000 CB 0,3% 0,5% US Caucasians (60) United States 7 HapMap data3 CB 0% 0% Swedes (50) Sweden 7 Patin et al. 20062 CB 0% 0% Saami (48) Finland 7 2 CB 0% 0% M+CB 0,1% 0,5% CB 0,9% 1,8% M+CB 0% 0% M+CB 0% 0% Patin et al. 2006 11 Germans (844) Germany 7 Cascorbi et al. 1995 Germans (223) Germany 6 Schnakenberg et al. 200012 Czechs (27) Czech Republic 6 Polish (248) Poland 7 Luca et al. 2008 4 13 Mrozikiewicz et al. 1996 Table S1 (Continued) Population (No. of individuals) Ashkenazi Jews (40) Sampling location - No. of screened Reference a variants 7 Patin et al. 20062 14 Haplotyping Classification error rate Classification error rate methodb for two phenotypesc for three phenotypesd CB 0% 0% CB 0% 0% 0% 7,1% Romanians (140) Romania 6 Rabstein et al. 2006 Mordvins (14) Mordovia 6 Luca et al. 20084 M+CB Russians (290) Russia 7 Gaikovitch et al. 2003 M+CB 0% 0% Perm Russians (12) Russia 6 Luca et al. 20084 M+CB 0% 8,3% Khanty&Mansi (14) Russia 6 Luca et al. 20084 M+CB 0% 0% Turkmen (50) Uzbekistan 7 Patin et al. 20062 CB 0% 0% Kyrgyz (290) Kyrgyzstan 6 Rabstein et al. 200614 CB 0% 0% Central/South Asia (556) 2 Gujarati (50) India 7 Patin et al. 2006 CB Dravidians (166) India 7 Anitha and Banerjee 200315 M Han Chinese (112) China 7 Guo et al. 200416 CB 0% Han Chinese (45) China 7 HapMap data3 CB 0% 0% Chinese (44) China 7 Patin et al. 20062 CB 0% 2,3% Japanese (144) Japan 7 Tanaka et al. 200217 CB 0% 0,7% 0% 0% 1,2% 10,2% 1,8% East Asia (2,134) 18 Japanese (172) Japan 6 Deguchi et al. 2005 CB 0% 0% Japanese (44) Japan 7 HapMap data3 CB 0% 0% Koreans (288) Korea 7 Lee et al. 200319 CB 0% 2,1% Koreans (1,000) Korea 6 Lee et al. 200220 0,2% 2,6% Thai (44) Thailand 7 M CB 0% 0% 0% 2 Patin et al. 2006 21 Indonesians (212) Indonesia 7 Yuliwulandari et al. 2007 CE 0% Yakuts (16) Siberia 6 Luca et al. 20084 M+CB 0% 0% Chukchee (13) Siberia 6 Luca et al. 20084 M+CB 0% 7,7% Embera (136) Panama 6 Jorge-Nebert et al. 200222 CC 0% 2,2% Ngawbe (105) Panama 6 Jorge-Nebert et al. 200222 CC 0% 2,9% Nicaraguans (137) Nicaragua 7 M 0% 1,5% America (378) 23 Martinez et al. 1998 a All population samples were genotyped for the seven common SNPs of the NAT2 gene (191G>A, 282C>T, 341T>C, 481C>T, 590G>A, 803A>G, and 857G>A), except some non-African samples where the SNP 191G>A was omitted since this is monomorphic in non-African populations. b In each sample, NAT2 haplotypes were reconstructed using either molecular techniques (M), computational algorithms (C), or a combination of both approaches (M+C; some studies indeed limited the application of molecular haplotyping to particular cases as those where an alternative linkage pattern of mutations would have led to a change in phenotype). CB, CE and CC refer to the computational method used (when applicable): the Bayesian algorithm implemented in PHASE24, the EM algorithm25 implemented in Haploview26, and the Clark’s method27 based on maximum parsimony, respectively. c Classification error rate of NAT2PRED when two phenotypic classes were considered: slow and other acetylators (intermediate and rapid acetylators pooled together). d Classification error rate of NAT2PRED when three phenotypic classes were considered: slow, intermediate and rapid acetylators. References 1. 2. 3. 4. 5. 6. 7. 8. 9. Loktionov A, Moore W, Spencer SP, Vorster H, Nell T, O'Neill IK, Bingham SA, Cummings JH. Differences in N-acetylation genotypes between Caucasians and Black South Africans: implications for cancer prevention. Cancer Detect. Prev. 26, 15-22 (2002). Patin E, Barreiro LB, Sabeti PC, Austerlitz F, Luca F, Sajantila A, Behar DM, Semino O, Sakuntabhai A, Guiso N, Gicquel B, McElreavey K, Harding RM, Heyer E, Quintana-Murci L: Deciphering the ancient and complex evolutionary history of human arylamine N-acetyltransferase genes. Am. J. Hum. Genet. 78, 423-436 (2006). The International HapMap Consortium. The International HapMap Project. Nature 426, 789-796 (2003). Luca F, Bubba G, Basile M, Brdicka R, Michalodimitrakis E, Rickards O, Vershubsky G, Quintana-Murci L, Kozlov AI, Novelletto A. Multiple advantageous amino acid variants in the NAT2 gene in human populations. PLoS ONE 3, e3136 (2008). Delomenie C, Sica L, Grant DM, Krishnamoorthy R, Dupret JM. Genotyping of the polymorphic N-acetyltransferase (NAT2*) gene locus in two native African populations. Pharmacogenetics 6, 177-185 (1996). Sabbagh A, Langaney A, Darlu P, Gérard N, Krishnamoorthy R, Poloni ES. Worldwide distribution of NAT2 diversity: implications for NAT2 evolutionary history. BMC Genet. 9, 21 (2008). Al-Yahyaee S, Gaffar U, Al-Ameri MM, Qureshi M, Zadjali F, Ali BH, Bayoumi R. N-acetyltransferase polymorphism among northern Sudanese. Hum. Biol. 79, 445-452 (2007). Agúndez JA, Golka K, Martínez C, Selinski S, Blaszkewicz M, García-Martín E. Unraveling ambiguous NAT2 genotyping data. Clin. Chem. 54, 13901394 (2008). Aynacioglu AS, Cascorbi I, Mrozikiewicz PM, Roots I. Arylamine N-acetyltransferase (NAT2) genotypes in a Turkish population. Pharmacogenetics 7, 327-331 (1997). 10. 11. 12. 13. 14. 15. 16. 17. 18. 19. 20. 21. 22. 23. 24. 25. 26. 27. Deitz AC, Zheng W, Leff MA, Gross M, Wen WQ, Doll MA, Xiao GH, Folsom AR, Hein DW. N-Acetyltransferase-2 genetic polymorphism, welldone meat intake, and breast cancer risk among postmenopausal women. Cancer Epidemiol. Biomarkers Prev. 9, 905-910 (2000). Cascorbi I, Drakoulis N, Brockmoller J, Maurer A, Sperling K, Roots I. Arylamine N-acetyltransferase (NAT2) mutations and their allelic linkage in unrelated Caucasian individuals: correlation with phenotypic activity. Am. J. Hum. Genet. 57, 581-592 (1995). Schnakenberg E, Lustig M, Breuer R, Werdin R, Hubotter R, Dreikorn K, Schloot W. Gender-specific effects of NAT2 and GSTM1 in bladder cancer. Clin. Genet. 57, 270-277 (2000). Mrozikiewicz PM, Cascorbi I, Brockmoller J, Roots I. Determination and allelic allocation of seven nucleotide transitions within the arylamine Nacetyltransferase gene in the Polish population. Clin. Pharmacol. Ther. 59, 376-382 (1996). Rabstein S, Unfried K, Ranft U, Illig T, Kolz M, Rihs HP, Mambetova C, Vlad M, Bruning T, Pesch B. Variation of the N-acetyltransferase 2 gene in a Romanian and a Kyrgyz population. Cancer Epidemiol. Biomarkers Prev. 15, 138-141 (2006). Anitha A, Banerjee M. Arylamine N-acetyltransferase 2 polymorphism in the ethnic populations of South India. Int. J. Mol. Med. 11, 125-131 (2003). Guo WC, Lin GF, Zha YL, Lou KJ, Ma QW, Shen JH. N-Acetyltransferase 2 gene polymorphism in a group of senile dementia patients in Shanghai suburb. Acta Pharmacol. Sin. 25, 1112-1117 (2004). Tanaka E, Taniguchi A, Urano W, Nakajima H, Matsuda Y, Kitamura Y, Saito M, Yamanaka H, Saito T, Kamatani N. Adverse effects of sulfasalazine in patients with rheumatoid arthritis are associated with diplotype configuration at the N-acetyltransferase 2 gene. J. Rheumatol. 29, 2492-2499 (2002). Deguchi M, Yoshida S, Kennedy S, Ohara N, Motoyama S, Maruo T. Lack of association between endometriosis and N-acetyl transferase 1 (NAT1) and 2 (NAT2) polymorphisms in a Japanese population. J. Soc. Gynecol. Investig. 12, 208-213 (2005). Lee KM, Park SK, Kim SU, Doll MA, Yoo KY, Ahn SH, Noh DY, Hirvonen A, Hein DW, Kang D. N-acetyltransferase (NAT1, NAT2) and glutathione S-transferase (GSTM1, GSTT1) polymorphisms in breast cancer. Cancer Lett. 196, 179-186 (2003). Lee SY, Lee KA, Ki CS, Kwon OJ, Kim HJ, Chung MP, Suh GY, Kim JW. Complete sequencing of a genetic polymorphism in NAT2 in the Korean population. Clin. Chem. 48, 775-777 (2002). Yuliwulandari R, Sachrowardi Q, Nishida N, Takasu M, Batubara L, Susmiarsih TP, Rochani JT, Wikaningrum R, Miyashita R, Miyagawa T, Sofro AS, Tokunaga K. Polymorphisms of promoter and coding regions of the arylamine N-acetyltransferase 2 (NAT2) gene in the Indonesian population: proposal for a new nomenclature. J. Hum. Genet. 53, 201-209 (2008). Jorge-Nebert LF, Eichelbaum M, Griese EU, Inaba T, Arias TD. Analysis of six SNPs of NAT2 in Ngawbe and Embera Amerindians of Panama and determination of the Embera acetylation phenotype using caffeine. Pharmacogenetics 12, 39-48 (2002). Martinez C, Agundez JA, Olivera M, Llerena A, Ramirez R, Hernandez M, Benitez J. Influence of genetic admixture on polymorphisms of drugmetabolizing enzymes: analyses of mutations on NAT2 and C gamma P2E1 genes in a mixed Hispanic population. Clin. Pharmacol. Ther. 63, 623-628 (1998). Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 73, 11621169 (2003). Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol. Biol. Evol. 12, 921-927 (1995). Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005). Clark AG. Inference of haplotypes from PCR-amplified samples of diploid populations. Mol. Biol. Evol. 7, 111-122 (1990).