Supplementary Materials
advertisement

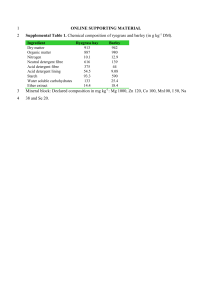
Supplementary Materials Materials and Methods Analysis of terminal restriction fragment length polymorphism data The abundance of individual terminal restriction fragments (TRFs) was calculated as previously outlined (Clement et al., 1998; Enright et al., 2007) except that TRFs representing <5% of the total area of respective restriction patterns were excluded from further analyses. Terminal restriction fragment length polymorphism (TRFLP) analysis could subsequently be successfully applied to monitor the fate of specific clones throughout the trial, taking into consideration that discrepancies of ± 2 to 3 base-pairs (bp) have to be taken into account when comparing predicted and measured TRFs (Liu et al., 1997; Kitts, 2001; Lueders and Friedrich, 2003). The identity of TRFs was assigned through (i) performing in silico enzyme restrictions of clone library sequences (ii) the TAP-TRFLP tool of the Ribosomal Database Project database (Marsh et al., 2000) and (iii) an in-house database of simulated archaeal and bacterial in silico restrictions (Collins et al., 2003; McHugh et al., 2003; McHugh et al., 2004; Enright et al., 2007, 2009). TRFLP data analysis was conducted either qualitatively, by creating binary matrices whereby the presence (‘1’) or absence (‘0’) of individual TRFs was scored, or semi-quantitatively, by calculating the relative abundance of TRFs normalised by the total area of the respective TRF patterns. Cluster analysis was performed using those matrices and a Multivariant Statistical Package (MVSP; Kovach, 1999). Dendrograms were constructed using unweighted pair-group methods using arithmetic averages (UPGMA) analysis and Jaccard distance measure. Ordination by non-metric multidimensional scaling (NMS; Kruskal, 1964) was also performed based on the presence/absence matrices to track the 1 temporal changes in community structure. NMS condenses a data set to a point on the resulting plot without loss of information so that the communities with similar TRFLP profiles will be plotted close together (McCune and Grace, 2002). In this study, the NMS ordination was carried out based on Sorensen (Bray-Curtis) distance measure with PC-ORD software (version 5.0; Grandin, 2006). Quantitative polymerase chain reaction analysis Quantitative polymerase chain reaction amplifications were performed in a two-step thermal cycling: pre-denaturation for 10 min at 94○C, followed by 40 cycles of 10 s at 94○C and 30 s at 60○C. All templates were analysed in triplicate. The quantification standard curves were constructed as previously described (Yu et al., 2006) using representative strains of the target groups: Methanospirillum hungatei JF1 (DSM 864) and Methanomicrobium mobile BP (DSM 1539) for MMB set and Methanosaeta concilli GP6 (DSM 3671) for Mst-set. Three custom recombinant plasmids containing the full-length 16S rRNA gene sequences of the representative strains were used as the templates for constructing standard curves. For MMB-set, the equimolar mixture of its two different standard plasmids was used. The mass concentrations of plasmids were measured in duplicate using a Qubit system (Invitrogen, U.S.A.) and converted to their copy concentrations (Yu et al., 2006). A 10-fold dilution series of 101 to 109 copies l-1 was generated for each standard solution and analysed in triplicate by qPCR. The threshold cycle values (CT) determined were plotted against the logarithm of their template copy concentrations. The 16S rRNA gene copy concentrations of target groups were determined against the corresponding standard curves within the linear range (r2 >0.995). 2 Table SM1. Archaeal clone library analysis; In silico TRFLP restrictions of clonal sequences. Library Accession No. Coverage (%) Lineage1 Seed DQ679927 DQ679928 DQ679929 DQ679930 DQ679931 DQ679932 DQ679933 13 3 3 10 7 13 51 Uncultured Crenarchaeota Uncultured Crenarchaeota Methanosaeta Methanosaeta Methanomethylovorans Methanosaeta Methanosaeta Sequence Homology TRFLP in silico restriction length3 (%)2 HhaI-forward HhaI-reverse 99 n.d. 231 99 n.d. 231 99 n.d. 577 99 n.d. 582 97 175 n.d. 98 193 n.d. 99 193 n.d. Day 1,228 FJ347527 FJ347528 FJ347529 FJ347530 FJ347531 FJ347532 43 19 30 4 3 1 Methanosaeta Methanocorpusculum Methanocorpusculum Methanocorpusculum Methanosaeta Methanocorpusculum 99 97 99 99 94 98 1 Based on Ribosomal Database Project classification. Based on National Centre for Biomedical Information classification. 3 In base-pairs. n.d. - not determined. 2 3 195 326 327 326 193 327 581 122 122 122 582 364 Figure SM1. Non metric multidimensional scaling (NMS) plot for the TRFLP spectra derived from (a) bacterial and (b) archaeal HhaI-reverse primer profiles. Percentage of variance for each axis is given in parenthesis. (a) (b) 4 Figure SM2. Cluster analysis (UPGMA) of bacterial terminal restriction fragments resolved using (a) HhaI-reverse and (b) HhaI-forward primer profiles. (a) (b) 5 Figure SM3. Cluster analysis (UPGMA) of archaeal terminal restriction fragments resolved using (a) HhaI-reverse and (b) HhaI-forward primer profiles. (a) (b) 6 References Clement BG, Kehl LE, DeBord KL, Kitts CL (1998). Terminal restriction fragment patterns (TRFPs), a rapid, PCR-based method for the comparison of complex bacterial communities. J Microbiol Methods 31: 135-142. Collins G, Woods A, McHugh S, Carton MW, O'Flaherty V (2003). Microbial community structure and methanogenic activity during start-up of psychrophilic anaerobic digesters treating synthetic industrial wastewaters. FEMS Microbiol Ecol 46: 159-170. Enright A-M, Collins G, O'Flaherty V (2007). Temporal microbial diversity changes in solvent-degrading anaerobic granular sludge from low-temperature (15oC) wastewater treatment bioreactors. Syst Appl Microbiol 30: 471-482. Enright A-M, McGrath V, Gill D, Collins G, O'Flaherty V (2009). Effect of seed sludge and operation conditions on performance and archaeal community structure of low-temperature anaerobic solvent-degrading bioreactors. Syst Appl Microbiol 32: 65-79. Grandin U (2006) PC-ORD version 5: A user-friendly toolbox for ecologists. J Veg Sci 17: 843-844. Kitts CL (2001). Terminal restriction fragment patterns: a tool for comparing microbial communities and assessing community dynamics. Current Issues in Intestinal Microbiology 2: 17-25. Kovach WL (1999) MVSP – a multivariate statistical package for windows, version 3.1. Kovach Computing Services. Wales, UK. Kruskal JB (1964). Nonmetric multidimensional-scaling - A numerical-method. Psychometrika 29: 115-129. Liu WT, Marsh TL, Cheng H, Forney LJ (1997). Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol 63: 4516-4522. Lueders T, Friedrich MW (2003). Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl Environ Microbiol 69: 320-326. Marsh TL, Saxman P, Cole J, Tiedje J (2000). Terminal restriction fragment length polymorphism analysis program, a web-based research tool for microbial community analysis. Appl Environ Microbiol 66: 3616-3620. McCune B & Grace JB (2002) Analysis of ecological communities. MJM Software Design, Gleneden Beach, OR, U.S.A. McHugh S, Carton M, Collins G, O'Flaherty V (2004). Reactor performance and microbial community dynamics during anaerobic biological treatment of wastewaters at 16-37○C. FEMS Microbiol Ecol 48: 369-378. McHugh S, Carton M, Mahony T, O'Flaherty V (2003). Methanogenic population structure in a variety of anaerobic bioreactors. FEMS Microbiol Lett 219: 297304. Yu Y, Kim J, Hwang S (2006). Use of real-time PCR for group-specific quantification of aceticlastic methanogens in anaerobic processes: Population dynamics and community structures. Biotechnol Bioeng 93: 424-433. 7