Author template for journal articles
advertisement
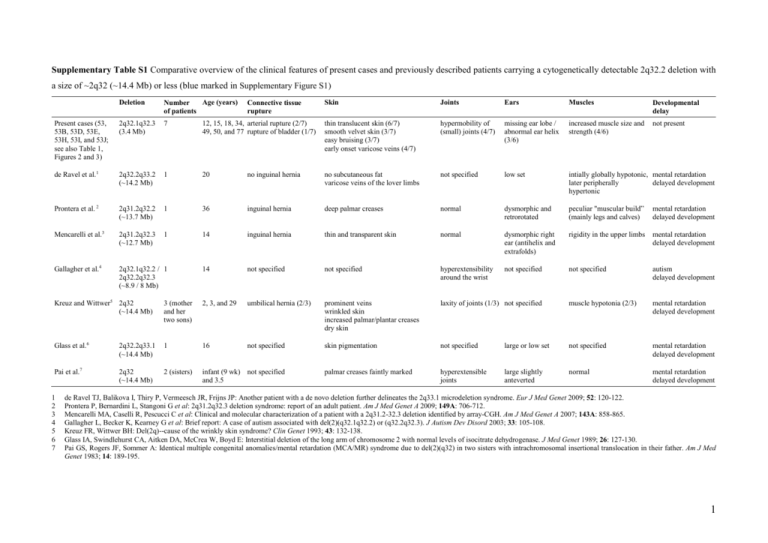
Supplementary Table S1 Comparative overview of the clinical features of present cases and previously described patients carrying a cytogenetically detectable 2q32.2 deletion with a size of ~2q32 (~14.4 Mb) or less (blue marked in Supplementary Figure S1) Deletion Age (years) Number of patients Skin Joints Ears Muscles Developmental delay Present cases (53, 53B, 53D, 53E, 53H, 53I, and 53J; see also Table 1, Figures 2 and 3) 2q32.1q32.3 (3.4 Mb) 7 12, 15, 18, 34, arterial rupture (2/7) 49, 50, and 77 rupture of bladder (1/7) thin translucent skin (6/7) smooth velvet skin (3/7) easy bruising (3/7) early onset varicose veins (4/7) hypermobility of (small) joints (4/7) missing ear lobe / abnormal ear helix (3/6) increased muscle size and strength (4/6) not present de Ravel et al.1 2q32.2q33.2 (~14.2 Mb) 1 20 no inguinal hernia no subcutaneous fat varicose veins of the lover limbs not specified low set intially globally hypotonic, mental retardation later peripherally delayed development hypertonic Prontera et al. 2 2q31.2q32.2 (~13.7 Mb) 1 36 inguinal hernia deep palmar creases normal dysmorphic and retrorotated peculiar "muscular build” (mainly legs and calves) mental retardation delayed development Mencarelli et al.3 2q31.2q32.3 (~12.7 Mb) 1 14 inguinal hernia thin and transparent skin normal dysmorphic right ear (antihelix and extrafolds) rigidity in the upper limbs mental retardation delayed development Gallagher et al.4 2q32.1q32.2 / 1 2q32.2q32.3 (~8.9 / 8 Mb) 14 not specified not specified hyperextensibility around the wrist not specified not specified autism delayed development Connective tissue rupture Kreuz and Wittwer5 2q32 (~14.4 Mb) 3 (mother and her two sons) 2, 3, and 29 umbilical hernia (2/3) prominent veins wrinkled skin increased palmar/plantar creases dry skin laxity of joints (1/3) not specified muscle hypotonia (2/3) mental retardation delayed development Glass et al.6 2q32.2q33.1 (~14.4 Mb) 1 16 not specified skin pigmentation not specified large or low set not specified mental retardation delayed development Pai et al.7 2q32 (~14.4 Mb) 2 (sisters) infant (9 wk) not specified and 3.5 palmar creases faintly marked hyperextensible joints large slightly anteverted normal mental retardation delayed development 1 2 3 4 5 6 7 de Ravel TJ, Balikova I, Thiry P, Vermeesch JR, Frijns JP: Another patient with a de novo deletion further delineates the 2q33.1 microdeletion syndrome. Eur J Med Genet 2009; 52: 120-122. Prontera P, Bernardini L, Stangoni G et al: 2q31.2q32.3 deletion syndrome: report of an adult patient. Am J Med Genet A 2009; 149A: 706-712. Mencarelli MA, Caselli R, Pescucci C et al: Clinical and molecular characterization of a patient with a 2q31.2-32.3 deletion identified by array-CGH. Am J Med Genet A 2007; 143A: 858-865. Gallagher L, Becker K, Kearney G et al: Brief report: A case of autism associated with del(2)(q32.1q32.2) or (q32.2q32.3). J Autism Dev Disord 2003; 33: 105-108. Kreuz FR, Wittwer BH: Del(2q)--cause of the wrinkly skin syndrome? Clin Genet 1993; 43: 132-138. Glass IA, Swindlehurst CA, Aitken DA, McCrea W, Boyd E: Interstitial deletion of the long arm of chromosome 2 with normal levels of isocitrate dehydrogenase. J Med Genet 1989; 26: 127-130. Pai GS, Rogers JF, Sommer A: Identical multiple congenital anomalies/mental retardation (MCA/MR) syndrome due to del(2)(q32) in two sisters with intrachromosomal insertional translocation in their father. Am J Med Genet 1983; 14: 189-195. 1 Supplementary Table S2 Laboratory blood parameters in deletion-carrier grandmother (53D), father (53E), son (53J), and daughter (53I) as well as in deletion-noncarrier mother (53M, as control subject) Parameter 53M (1962) Reference range Result Unit Clinical chemistry Total bilirubin 53D (1932) Reference range Result Unit 6 μmol/L <17 7 μmol/L <21 Creatinine Creatine kinase C-reactive protein 63 μmol/L 44-80 107 U/L <167 <1 mg/L <5 76 μmol/L 44-80 163 U/L <167 <1 mg/L <5 Serum iron Ferritin Transferrin Transferrin saturation Soluble transferrin receptor 9.3 10 38 0.12 4.22 μmol/L 7.0-26.0 μg/L 10-150 μmol/L 25-50 0.15-0.50 mg/L 1.9-4.4 Hematology Hemoglobin Hematocrit Red blood cell count MCV MCH MCHC Microcytes Macrocytes Hypochromic RBC Hyperchromic RBC Platelets Left shift White blood cell count 12.2 36.0 4.09 88.1 29.9 34.0 0.7 0.3 1.4 0.5 297 0 4.48 g/dl % 106/μl fl pg g/dl % % % % 103/μl 2.46 0.31 0.11 0.05 1.44 2.6 3 10 /μl 103/μl 103/μl 103/μl 103/μl % 1.40-8.00 0.16-0.95 0.00-0.70 0.00-0.15 1.50-4.00 0.0-4.0 0.74 0.1 <8 negative 1 g/L g/L IU/ml 0.9-1.8 0.1-0.4 <20 <1:100 <30 Neutrophils Monocytes Eosinophils Basophils Lymphocytes LUC (non-classifiable lymphoblast cells) Immunological assays Complement C3c Complement C4 Rheumatoid factor ANA Anti-CCP 103/μl U/ml 11.7-15.3 35-46 3.9-5.2 80-100 26-34 31-36 0-2.0 0-2.0 0-2.0 0-2.0 143-400 no (0) 3.0-9.6 12.7 85 35 0.18 1.17 53E (1960) Reference range Result Unit 9 μmol/L <17 76 μmol/L 62-106 98 U/L <190 <1 mg/L <5 53J (1990) Reference range Result Unit 11 μmol/L <17 80 μmol/L 62-106 134 U/L <190 <1 mg/L <5 53I (1993) Reference range Result Unit 10 μmol/L <17 53 μmol/L 44-80 78 U/L <167 <1 mg/L <5 μmol/L 7.0-26.0 μg/L 30-400 μmol/L 25-50 0.15-0.50 mg/L 0.76-1.76 15 108 27 0.28 2.68 μmol/L 11.0-28.0 μg/L 30-400 μmol/L 25-50 0.20-0.55 mg/L 2.2-5.0 23.4 40 31 0.38 2.83 μmol/L 11.0-28.0 μg/L 30-400 μmol/L 25-50 0.20-0.55 mg/L 2.2-5.0 28.4 49 36 0.39 2.88 μmol/L 7.0-26.0 μg/L 20-200 μmol/L 25-50 0.15-0.50 mg/L 1.9-4.4 not performed not performed not performed not performed not performed not performed not performed not performed not performed not performed not performed not performed not performed 14.7 43.3 4.95 87.3 29.6 33.9 0.5 0.2 0.8 0.8 207 0 7.11 g/dl % 106/μl fl pg g/dl % % % % 103/μl 15.2 43.4 5.22 83.0 29.0 35.0 1.1 0.1 0.5 1.6 182 0 5.88 g/dl % 106/μl fl pg g/dl % % % % 103/μl 13.4-17.0 40-50 4.2-5.7 80-100 26-34 31-36 0-2.0 0-2.0 0-2.0 0-2.0 143-400 no (0) 3.0-9.6 13.5 40.2 4.66 86.2 29.0 33.7 1.0 0.1 2.8 0.4 265 0 5.99 g/dl % 106/μl fl pg g/dl % % % % 103/μl 103/μl 11.7-15.3 35-46 3.9-5.2 80-100 26-34 31-36 0-2.0 0-2.0 0-2.0 0-2.0 143-400 no (0) 3.0-9.6 not performed not performed not performed not performed not performed not performed 4.51 0.55 0.25 0.03 1.60 2.3 3 10 /μl 103/μl 103/μl 103/μl 103/μl % 1.40-8.00 0.16-0.95 0.00-0.70 0.00-0.15 1.50-4.00 0.0-4.0 3.30 0.41 0.46 0.05 1.54 2.0 3 10 /μl 103/μl 103/μl 103/μl 103/μl % 1.40-8.00 0.16-0.95 0.00-0.70 0.00-0.15 1.50-4.00 0.0-4.0 3.25 0.40 0.37 0.05 1.77 2.6 103/μl 103/μl 103/μl 103/μl 103/μl % 1.40-8.00 0.16-0.95 0.00-0.70 0.00-0.15 1.50-4.00 0.0-4.0 not performed not performed not performed not performed not performed 0.83 0.18 <8 negative 0 g/L g/L IU/ml 0.9-1.8 0.1-0.4 <20 <1:100 <30 0.85 0.09 <8 negative 0 g/L g/L IU/ml 0.9-1.8 0.1-0.4 <20 <1:100 <30 0.89 0.14 <8 negative 0 g/L g/L IU/ml 0.9-1.8 0.1-0.4 <20 <1:100 <30 103/μl U/ml 13.4-17.0 40-50 4.2-5.7 80-100 26-34 31-36 0-2.0 0-2.0 0-2.0 0-2.0 143-400 no (0) 3.0-9.6 103/μl U/ml U/ml Dietary habits: 53D and 53E are no vegetarians, whereas 53I and 53J are vegetarians for the last ~1.5 years and ~6 months, respectively, but both eat fish. Due to the lack of long-term low iron intake, dietary habits of these deletion-carriers may not influence iron homeostasis. However, the effect of dietary habits on HFE4 has not been investigated. Clinical chemistry: Venous plasma and serum samples were obtained by venous puncture and subsequent centrifugation of the blood samples. The concentrations of creatinine, total bilirubin, C-reactive protein (CRP), serum iron, and creatine kinase were assayed on a Roche/Hitachi Modular System P (Roche Diagnostics, Rotkreuz, Switzerland) according to the manufacturer’s specifications and using proprietary reagents. The ferritin immunoassay was performed on the Modular Analytics E170 analyzer (Roche Diagnostics, Rotkreuz, Switzerland). Transferrin was measured on a COBAS Integra analyzer (Roche Diagnostics, Rotkreuz, Switzerland). Soluble transferrin receptor (sTfR) was analyzed using a nephelometric technique (Dade Behring, Marburg, Germany). Value of slightly elevated serum iron in 53I was not further investigated. Hematology: Blood counts were analyzed using an Advia 2120 automated hematology system (Siemens Healthcare Diagnostics, Eschborn, Germany). Immunological assays: Rheumatoid factor (RF) and complement factors C4 and C3c were determined on a Dade Behring nephelometer BNII (Dade Behring, Düdingen, Switzerland) according to the manufacturer’s instructions (low C3c values are most likely due to immediate analysis; i.e. there was very short pre-analytical delay). Antinuclear antibodies (ANA) were determined by indirect immunofluorescence microscopy using Hep2-cells and rat tissues (Euroimmun Basis Profil 3B FB1805-2005-3). ANA titers less than 1:100 were considered negative. Antibodies to cyclic citrullinated peptide (Anti-CCP) were measured by Immunoscan RA anti-CCP ELISA (Euro-Diagnostica, Malmö, Sweden). Immunological assays were performed to assess the impact of the partial deletion of STAT4 (Figure 1), which has been associated with rheumatoid arthritis (RA, MIM #180300) and systemic lupus erythematosus (SLE, MIM #152700) (Supplementary Table S1). RF and Anti-CCP were analyzed regarding RA and ANA regarding SLE (anti-dsDNA and antiSm antibodies were not analyzed because ANA was negative). The analyzed parameters were in all three deletion-carriers within the normal range, providing no evidence for RA and SLE. This lack of association of the partial deletion of STAT4 with RA and SLE is not inconsistent with current knowledge. However, RA and SLE cannot be completely excluded by these immunological assays. 2 Supplementary Table S3 Known copy number variations (CNVs) larger than 1 kb within the deletion of 3.4 Mb identified in this study Namea Type Start position End position Size (bp) Detection rateb Known genes Reference 6015 Loss 188,360,896 188,363,219 2,324 1/36 --Mills et al.1 1042 Loss 189,277,306 189,287,290 9,985 1/30 --Conrad et al.2 32389 Loss 189,278,292 189,286,640 8,349 2/30 --Perry et al.3 37938 Loss 189,276,201 189,287,574 11,374 17/270 --McCarroll et al.4 43660 Loss 189,438,791 189,448,384 9,593 1/1 --Wang et al.5 0029 Gain 190,008,980 190,176,870 167,891 2/39 WDR75, SLC40A1 Iafrate et al.6 6016 Loss 190,434,734 190,437,628 2,895 1/36 PMS1 Mills et al.1 7242 Gain 191,027,036 191,073,955 46,920 2/50 MFSD6 De Smith et al.7 35774 Gain 191,169,323 191,199,654 30,332 1/1 --Kidd et al.8 22737 Loss 191,382,600 191,389,807 7,208 1/2 --Korbel et al.9 a According to the Database of Genomic Variants (DGV, http://projects.tcag.ca/variation, version May, 2009) b Number of individuals harbouring the corresponding CNV in relation to the total number of individuals analyzed 1 Mills RE, Luttig CT, Larkins CE et al: An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res 2006; 16: 1182-1190. 2 Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK: A high-resolution survey of deletion polymorphism in the human genome. Nat Genet 2006; 38: 75-81. 3 Perry GH, Ben-Dor A, Tsalenko A et al: The fine-scale and complex architecture of human copy-number variation. Am J Hum Genet 2008; 82: 685-695. 4 McCarroll SA, Kuruvilla FG, Korn JM et al: Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet 2008; 40: 1166-1174. 5 Wang J, Wang W, Li R et al: The diploid genome sequence of an Asian individual. Nature 2008; 456: 60-65. 6 Iafrate AJ, Feuk L, Rivera MN et al: Detection of large-scale variation in the human genome. Nat Genet 2004; 36: 949-951. 7 De Smith AJ, Tsalenko A, Sampas N et al: Array CGH analysis of copy number variation identifies 1284 new genes variant in healthy white males: implications for association studies of complex diseases. Hum Mol Genet 2007; 16: 2783-2794. 8 Kidd JM, Cooper GM, Donahue WF et al: Mapping and sequencing of structural variation from eight human genomes. Nature 2008; 453: 56-64. 9 Korbel JO, Urban AE, Affourtit JP et al: Paired-end mapping reveals extensive structural variation in the human genome. Science 2007; 318: 420-426. 3 Present case Rifai et al.1 de Ravel et al.2 starts at 2q24 starts between 2q24.2 and 2q24.3 Patient 2 Prontera et al.3 Gambrelle et al.4 Mencarelli et al.5 Bijlsma et al.6 Van Buggenhout et al.7 Van Buggenhout et al.7 Patient 3 may end at 2q35 Gallagher et Loscalzo et al.8 al.9 Kondo et al.10 may start at 2q24.3 Nixon et al.11 Kreuz and Wittwer12 Hiroyama et al.13 Palmer et al.14 Ramer et al.15 Glass et al.16 Ramer et al.17 Dallapiccola et al.18 Miyazaki et al.19 Sumi et al.20 Benson et al.21 Muneer et al.22 ends at 2q35 Markovic et al.23 Al-Awadi et al.24 Buchanan et al.25 Franceschini et al.26 Pai et al.27 Young et al.28 starts at 2q23 Shabtai et al.29 Taysi et al.30 Supplementary Figure S1 Overview of previously reported cases with cytogenetically detectable deletions affecting the region deleted in this study (Present case). Deletions larger than del(2)(q23q35) are not included because such aberrations affect much larger chromosomal regions and thus much more genes. Cases (deletions <15 Mb) compared in Supplementary Table S1 are indicated by blue colour. The maximal possible extent of the deletions is indicated (dashed lines). 1 Rifai L, Port-Lis M, Tabet AC et al: Ectodermal dysplasia-like syndrome with mental retardation due to contiguous gene deletion: further clinical and molecular delineation of del(2q32) syndrome. Am J Med Genet A 2010; 152A: 111-117. 2 de Ravel TJ, Balikova I, Thiry P, Vermeesch JR, Frijns JP: Another patient with a de novo deletion further delineates the 2q33.1 microdeletion syndrome. Eur J Med Genet 2009; 52: 120-122. 3 Prontera P, Bernardini L, Stangoni G et al: 2q31.2q32.3 deletion syndrome: report of an adult patient. Am J Med Genet A 2009; 149A: 706-712. 4 Gambrelle J, Till M, Lukusa B et al: Ocular anomalies associated with interstitial deletion of chromosome 2q31: case report and review. Ophthalmic Genet 2007; 28: 105-109. 5 Mencarelli MA, Caselli R, Pescucci C et al: Clinical and molecular characterization of a patient with a 2q31.2-32.3 deletion identified by array-CGH. Am J Med Genet A 2007; 143A: 858-865. 6 Bijlsma EK, Knegt AC, Bilardo CM, Goodman FR: Increased nuchal translucency and split-hand/foot malformation in a fetus with an interstitial deletion of chromosome 2q that removes the SHFM5 locus. Prenat Diagn 2005; 25: 39-44. 7 Van Buggenhout G, Van Ravenswaaij-Arts C, Mc Maas N et al: The del(2)(q32.2q33) deletion syndrome defined by clinical and molecular characterization of four patients. Eur J Med Genet 2005; 48: 276-289. 8 Loscalzo ML, Galczynski RL, Hamosh A, Summar M, Chinsky JM, Thomas GH: Interstitial deletion of chromosome 2q32-34 associated with multiple congenital anomalies and a urea cycle defect (CPS I deficiency). Am J Med Genet A 2004; 128A: 311-315. 9 Gallagher L, Becker K, Kearney G et al: Brief report: A case of autism associated with del(2)(q32.1q32.2) or (q32.2q32.3). J Autism Dev Disord 2003; 33: 105-108. 10 Kondo K, Kaga K, Ogawa Y, Fukushima Y: Temporal bone histopathologic findings in a case of interstitial deletion of the long arm of chromosome 2 [del(2) (q31q33)]. Int J Pediatr Otorhinolaryngol 1999; 48: 31-37. 11 Nixon J, Oldridge M, Wilkie AO, Smith K: Interstitial deletion of 2q associated with craniosynostosis, ocular coloboma, and limb abnormalities: cytogenetic and molecular investigation. Am J Med Genet 1997; 70: 324-327. 12 Kreuz FR, Wittwer BH: Del(2q)--cause of the wrinkly skin syndrome? Clin Genet 1993; 43: 132-138. 13 Hiroyama Y, Hatanaka H, Ikenoue T, Ishihara Y: Interstitial deletion of long arm of chromosome 2(q31q33). Acta Paediatr Jpn 1990; 32: 563-565. 14 Palmer CG, Heerema N, Bull M: Deletions in chromosome 2 and fragile sites. Am J Med Genet 1990; 36: 214-218. 15 Ramer JC, Mowrey PN, Robins DB, Ligato S, Towfighi J, Ladda RL: Five children with del (2)(q31q33) and one individual with dup (2)(q31q33) from a single family: review of brain, cardiac, and limb malformations. Am J Med Genet 1990; 37: 392-400. 16 Glass IA, Swindlehurst CA, Aitken DA, McCrea W, Boyd E: Interstitial deletion of the long arm of chromosome 2 with normal levels of isocitrate dehydrogenase. J Med Genet 1989; 26: 127-130. 17 Ramer JC, Ladda RL, Frankel CA, Beckford A: A review of phenotype-karyotype correlations in individuals with interstitial deletions of the long arm of chromosome 2. Am J Med Genet 1989; 32: 359-363. 18 Dallapiccola B, Novelli G, Giannotti A: Deletion 2q31.3----2q33.3: gene dosage effect of ribulose 5-phosphate 3-epimerase. Hum Genet 1988; 79: 92. 19 Miyazaki K, Yamanaka T, Ogasawara N: Interstitial deletion 2q32.1----q34 in a child with half normal activity of ribulose 5-phosphate 3-epimerase (RPE). J Med Genet 1988; 25: 850-851. 20 Sumi S, Obayashi M, Murakami M et al: Interstitial deletion of the long arm of chromosome 2: a dose study on isocitrate dehydrogenase 1. Jinrui Idengaku Zasshi 1988; 33: 461-467. 21 Benson K, Gordon M, Wassman ER, Tsi C: Interstitial deletion of the long arm of chromosome 2 in a malformed infant with karyotype 46,XX,del(2)(q31q33). Am J Med Genet 1986; 25: 405-411. 22 Muneer RS, Payne-Howell RM, Altshuler G, Rennert OM: An interesting case of partial monosomy 2q and a 3q:12q translocation. Pediatr Res 1986; 20: 269A. 23 Markovic S, Krstic M, Sulovic V, Radojkovic Z, Adzic S: Interstitial deletion of chromosome 2. J Med Genet 1985; 22: 154-155. 24 Al-Awadi SA, Farag TI, Naguib K et al: Interstitial deletion of the long arm of chromosome 2: del(2)(q31q33). J Med Genet 1983; 20: 464-465. 25 Buchanan PD, Rhodes RL, Stevenson CE, Jr.: Interstitial deletion 2q31 leads to q33. Am J Med Genet 1983; 15: 121-126. 26 Franceschini P, Cirillo Silengo M, Davi G, Bianco R, Biagioli M: Interstitial deletion of the long arm of chromosome 2 (q31q33) in a girl with multiple anomalies and mental retardation. Hum Genet 1983; 64: 98. 27 Pai GS, Rogers JF, Sommer A: Identical multiple congenital anomalies/mental retardation (MCA/MR) syndrome due to del(2)(q32) in two sisters with intrachromosomal insertional translocation in their father. Am J Med Genet 1983; 14: 189-195. 28 Young RS, Shapiro SD, Hansen KL, Hine LK, Rainosek DE, Guerra FA: Deletion 2q: two new cases with karyotypes 46,XY,del(2)(q31q33) and 46,XX,del(2)(q36). J Med Genet 1983; 20: 199-202. 29 Shabtai F, Klar D, Halbrecht I: Partial monosomy of chromosome 2. Delineable syndrome of deletion 2 (q23-q31). Ann Genet 1982; 25: 156-158. 30 Taysi K, Dengler DR, Jones LA, Heersma JR: Interstitial deletion of the long arm of chromosome 2: case report and review of literature. Ann Genet 1981; 24: 245-247. 4 1.5 0.5 0.0 COL3A1 Ex 51 COL3A1 Ex 32 COL3A1 Ex 17 COL3A1 Ex 29 COL3A1 Ex 20 COL3A1 Ex 43 COL3A1 Ex 11 COL3A1 Ex 2 COL3A1 Ex 5 1.0 COL3A1 Ex 1 1.0 Patient 53B COL3A1 Ex 1 Control Relative peak area Relative peak area 1.5 0.5 0.0 1 3 5 7 9 11 13 15 17 19 21 23 25 27 29 31 33 35 37 39 41 43 45 MLPA P155 fragment 1 3 5 7 9 11 13 15 17 19 21 23 25 27 29 31 33 35 37 39 41 43 45 MLPA P155 fragment Supplementary Figure S2 Result of MLPA analyses in patient 53B. The P155 kit contains 11 probes for COL3A1 (fragments 4, 9, 12, 15, 19, 21, 31, 35, 38, 40, and 44; red bars) and 14 control MLPA probes located on chromosomes 1, 5, 6, 7, 9, 11, 13, 17, 18, and 22 (fragments 1, 6, 7, 10, 14, 18, 20, 24, 27, 30, 36, 39, 42, and 45) as well as 14 probes for TNXB, two for CYP21A1P, and one probe for each of the genes C4B, LTA, BAK1, and CREBL1. The normal range of ±30% of the normalized relative peak areas is given by dotted lines. Note that in patient 53B all probes for COL3A1 showed a reduced relative peak area outside the normal range (for reduced peaks, the corresponding exons are denoted) and that there are two MLPA probes for COL3A1 exon 1. 5









