Enzyme substrate competition in a developing
advertisement

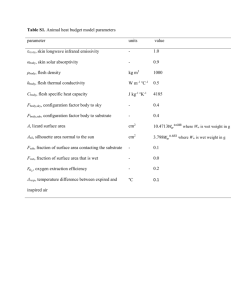
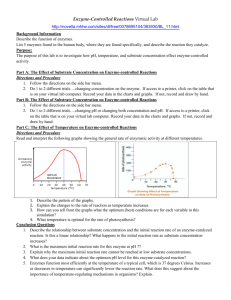
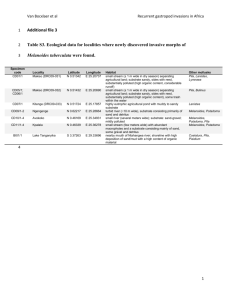
Enzyme substrate competition in a developing embryo Yoosik Kim*, Mathieu Coppey, and Stanislav Y. Shvartsman Department of Chemical Engineering and Lewis-Sigler Institute for Integrative Genomics, Princeton University, Princeton, NJ 08544, USA. Rona Grossman and Ze’ev Paroush Department of Developmental Biology and Cancer Research, Institute for Medical Research – Israel-Canada, The Hebrew University-Hadassah Medical School, PO Box 12272, Jerusalem 91120, Israe Leiore Ajuria and Gerardo Jiménez Institut de Biologia Molecular de Barcelona-CSIC and Institució Catalana de Recerca i Estudis Avançats, Parc Científic de Barcelona, Barcelona 08028, Spain. Session 3, Talk 1 (1:40 PM) Whereas certain enzymes exhibit remarkable substrate specificity, others are quite promiscuous. Among kinases, for example, D-fructokinase phosphorylates only one substrate (D-fructose), yet protein kinases can have hundreds of substrates [1]. A key question for such enzymes is whether their multiple substrates are processed in an ordered fashion or concurrently, in a regime where they compete for access to a common regulator. While cells have evolved a number of strategies for avoiding competitive effects to ensure ordered substrate processing [2], substrate competition can be beneficial; for example, for generating thresholds in cell cycle control [3]. Here we propose that competitive effects can also provide a mechanism for integration of patterning signals in embryonic development. Based on genetic and imaging experiments, we argue that in the early Drosophila embryo multiple substrates of the Mitogen Activated Protein Kinase (MAPK) compete for this enzyme. As a consequence, the signaling activity of MAPK becomes dependent on the levels of its substrates. The resulting substrate competition network provides a new mechanism for spatial integration of the maternal morphogen gradients [4]. Competitive effects may play a more general role in spatial regulation of signaling networks, where enzymes, such as MAPK, interact with their multiple regulators and targets. [1] Ubersax, J. A. and Ferrell, J. E., Nature Reviews Molecular Cell Biology 8, 530 (2007). [2] Sullivan, M. and Morgan, D. O. Nature Reviews Molecular Cell Biology 8, 894 (2007). [3] Bhattacharyya, R. P., Remenyi, A., Yeh, B. J., and Lim, W. A. Annual Review of Biochemistry 75, 655 (2006). [4] St Johnston, D. and Nusslein-Volhard, C. Cell 68 (2), 201 (1992).