Phylogenetic diversity and Chemical Response of Anammox
advertisement

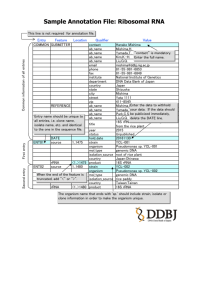
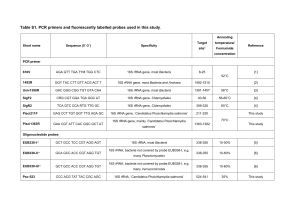
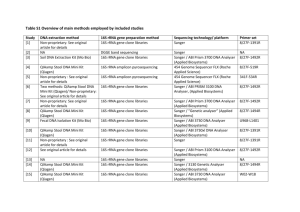
1 Supplementary materials 2 3 Mangrove Trees Affect the Community Structure and Distribution of Anammox 4 Bacteria at an Anthropogenic-Polluted Mangrove in the Pearl River Delta 5 Reflected by 16S rRNA and Hydrazine Oxidoreductase (HZO) Encoding Gene 6 Analyses 7 Meng Li1, Yi-Guo Hong1, 2, Hui-Luo Cao1 and Ji-Dong Gu1, 3* 8 9 10 11 12 13 Table S1. A comparison of clone libraries of 16S rRNA and hzo genes from the mangrove sediments a Homologous library X site P value for heterologous library Y site: S0-s S0-s 14 15 16 17 18 S1-s S1-l S2-s S2-l 0.001 (0.001) 0.001 (0.093) 0.001 (0.035) 0.001 (0.019) 0.001 (0.001) 0.013 (0.011) 0.001 (0.001) 0.018 (0.001) 0.026 (0.001) S1-s 0.001 (0.001) S1-l 0.001(0.001) 0.042 (0.001) S2-s 0.001 (0.001) 0.107 (0.204) 0.001 (0.245) S2-l 0.001 (0.001) 0.233 (0.839) 0.415 (0.998) 0.001 (0.006) 0.077 (0.99) a: In comparison between the homologous library X and the heterologous library Y, boldface P values indicate that the libraries of 16S rRNA and hzo genes clone are drawn from communities significantly difference (p<0.05). Numbers in parenthesis refer to the P value of comparing hzo gene clone libraries and those without parenthesis of 16S rRNA gene clone libraries. 19 Fig. S1 Rarefaction analysis of anammox bacterial communities based on 16S rRNA (A) and 20 hzo (B) gene sequences at the Mai Po Nature Reserve. DOTUR program was used with 2% 21 nucleotide or protein sequence variation as OTU determination. 22 8 S0-s S1-s S1-l S2-s S2-l 7 Number of OTUs observed 6 5 4 3 2 1 0 0 5 10 15 20 25 30 35 Number of clones sequenced 23 24 25 A 2 14 S0-s S1-s 12 S1-l Number of OTUs observed S2-s 10 S2-l 8 6 4 2 0 0 20 30 40 50 60 Number of clones sequenced 26 27 28 29 10 B S-Fig. 1 3