PB 150 Bioinformatics lab - College of Natural Resources
advertisement

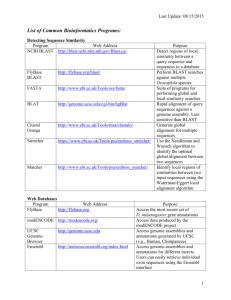
1 PB 150 Bioinformatics lab Objective: 1. An overview of genomes and plant biological resources databases. 2. Obtaining relevant scientific information starting from a keyword 3. Analyzing information and creating a database: Sequence alignment & Analysis at protein and nucleic acid level. Methodology NOTE: All the needed links for this lab, homework and more are listed below. * Access NCBI home page. http://www.ncbi.nlm.nih.gov/ From the main page, or ENTREZ, select [dropdown menu]: “Protein” * Use a keyword “tyrosine phosphatase” to search and retrieve sequences. Note be careful about spelling. * Look for mammalian and plant sequences [species name is in the title], leave the rest. Tip: Arabidopsis does not seem to have receptor-like tyrosine phosphatases, these sequence tend to result BLAST hits in MAPKs of Arabidopsis genome, you may want to gather as few of these as possible for your subsequent search. * Click on different sequences that fulfill these criteria: protein tyrosine phosphatase, from plant or mammal origin. * Select FASTA [dropdown menu, next to Display button] and click “Display”. Save the output in FASTA format to a text file [Copy and Paste is fine]. Note: FASTA format contains on the first line, behind a “>” information identifying each sequence, like accession number, species name, brief description of the sequence. Keep this information through the subsequent steps together with the actual protein sequence. * Access TAIR homepage, BLAST, http://arabidopsis.org/Blast/ select BLASTP [AA query, AA database; AGI Proteins (Protein)] from the dropdown menus, BLAST program and Datasets, respectively. Query sequence must be checked [it is default]. * Run a BLAST search with your candidate sequences. * Collect Arabidopsis sequences that share some homology. Look for hits that contain “phosphatase”, “myotubularin”, “PTEN”, “unknown protein”, “expressed protein” or “hypothetical protein”, in the title, save in FASTA format. 2 * Run each one individually at the Conserved Domain Database (integrates PFAM, SMART, etc) http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml Evaluate for relevance: - does it contain a DSPc or PTPc domain? A. Yes: it is a “good” sequence. B. No: that is the nature of BLAST: some of the hits may have only a distant homology, and/or homology in a domain that is different from the PTP/DSP catalytic domain. Collect all “good” sequences and save to a new text file. This is your small database (DB) that fulfills the following criteria: Contains - protein sequences of the - plant Arabidopsis, that are - protein tyrosine phosphatases. Question: Do you expect your database containing all Arabidopsis protein tyrosine phosphatases? Why? Homework: I. At protein level: i. Collect the gene accession numbers. Gene accession numbers are in the following format: Atxgyyyyy , where At is identifier for the organism: Arabidopsis thaliana, x is a chromosome number, which is from 1 to 5 for At, yyyyy is a gene specific number. Gene accession numbers may be available from the identifier string (1st line) of your sequences in FASTA format, so look first there. Unfortunately, it’s not necessarily the case. If not present, the Gene accession numbers can easily be retrieved as follow: Copy your sequence, one at the time, paste into TAIR BLAST search window. http://arabidopsis.org/Blast/ Select BLASTP and AGI Proteins (Protein) from the dropdown menus when working with protein sequence (the default is nucleotide, BLASTN). Run a BLAST search. The first hit(s) with E Value of 0.0 reveal(s) database entries with 100% identity over a certain stretch of sequence. This (these) must be the self-score(s) of your protein. The gene accession # is right after the hyperlinks. Save to your DB. [it can be saved in the FASTA > line] 3 ii. Start organizing your DB in a hierarchical manner: 1. numbers to each entry for your own reference, followed by gene accession numbers. 2. sequences in FASTA format corresponding to the #s you gave and the gene accession numbers. iii. Analyze your protein sequences for presence of: A. Chloroplast, Mitochondrion, Nucleus targeting signal peptides. B. Transmembrane domains. [choose appropriate links from the listed below in Specialty Tasks, Protein Specific, Subcellular Localization, or Transmembrane domains] Save your findings to your database in an organized, hierarchical manner. iv. Perform a ClustalW multiple sequence alignment with your sequence. http://www.ebi.ac.uk/clustalw/ ( or http://www.ch.embnet.org/software/ClustalW.html) Note: use FASTA format as in all your previous work. [scoring matrix BLOSUM is good, you may leave all the rest on default too, or try changing settings as “color”, etc] v. Build a phylogenetic tree with this program. It is automatically generated with the multiple sequence alignment. You can view the image of it at the bottom of you results page when using http://www.ebi.ac.uk/clustalw/ [this function may not work with all browsers!]. Alternatively, you will have to download /install software for viewing the tree (also called “dendrogram”). Note: Clustalw will display on the phylogenetic tree only the first string with limited number of characters as identifiers for your sequence(s). When running Clustalw, you may want to use your own reference numbers at the beginning of the FASTA “>” for easier identification/orientation in the tree’s homology relations. For example: >DSP1At3g1000gi…… ILOVETHISSEQENCEWORK…… Will display DSP1At3g1000gi to the branch with your ILOVETHISSEQENCEWORK…… sequence. II. At nucleic acid level: 1. Look for entries for “tyrosine phosphatase” in nucleic acid database. Following the same steps as during the demonstration, collect gene sequences for Arabidopsis protein tyrosine phosphatases. To verify if they are “real” tyrosine 4 phosphatases perform a BLAST search in the NT database [BLASTN, and AGI Genes (+ Introns, + UTRs) from dropdown menus]. 2. Crossreference with the gene accession numbers you already have to validate the entries. A. If you get new entries, not already present in your database, run their respective AA seq. in the CDD, you might just have found a new member of the PTP family. [AA sequence will be easy to find if you click on TAIR or TIGR hyperlinks, they are on the summary page of each sequence.] Email your results. A summary of your results will be posted temporarily on the unofficial web page of the course. In case of any questions, do not hesitate to ask Prof Luan, or Lubo. General useful links where and how to find (plant) science related information Literature: PubMed http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=PubMed ISI http://isi3.newisiknowledge.com/portal.cgi?DestApp=WOS&Func=Frame (address keeps changing, check it from here if link doesn’t work: http://isi3.newisiknowledge.com/portal.cgi ) Search in particular journal db and crossreferences therein i.e. http://www.plantcell.org/ http://www.plantphysiol.org/ Biochemical Pathways http://us.expasy.org/tools/pathways/ Enzyme nomenclature DB http://us.expasy.org/enzyme/ Protocols and procedures: Current Protocols http://www.mrw2.interscience.wiley.com/cponline/tserver.dll?command=doGetD oc&sUI=&database=CP&useScheme=WIS_Framed.Scheme&getDoc=cp_toc_fs. html Specialty tasks Sequences Retrieval ENTREZ @ NCBI (National Center for Biotechnology Information) 5 http://www3.ncbi.nlm.nih.gov/Entrez/ Sequences Analysis: Both Nucleic Acids & Protein Similarity and homology search: BLAST & FASTA concept HMM http://www.soe.ucsc.edu/research/compbio/HMMapps/T02-query.html Seq alignment 2 sequences http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html Multiple sequence analysis ClustalW http://www.ch.embnet.org/software/ClustalW.html T-Cofee http://www.es.embnet.org/Services/MolBio/t-coffee/ WebLogo http://weblogo.berkeley.edu/logo.cgi GCG registration required, if someone really interested in using it, contact the webmaster for arrangements. http://socrates.berkeley.edu:7029/gcg-bin/seqweb.cgi Phylogenetic trees ClustalW http://www.ch.embnet.org/software/ClustalW.html GCG http://socrates.berkeley.edu:7029/gcg-bin/seqweb.cgi DNA specific cis-acting regulatory DNA elements db-s (Promoter analysis) PLACE - Plant cis-acting regulatory DNA elements db PlantCARE - Plant cis-acting regulatory DNA elements db gene prediction: GENSCAN http://genes.mit.edu/GENSCAN.html GRAIL: http://compbio.ornl.gov/Grail-1.3/ Primer design: Primer3 http://www-genome.wi.mit.edu/cgibin/primer/primer3_www.cgi T-DNA mapping tool: http://signal.salk.edu/cgi-bin/tdnaexpress RNA specific: dbEST (Expressed Sequence Tags) db (NCBI) http://www.ncbi.nlm.nih.gov/dbEST/ or select EST from dropdown menu in BLAST search 6 Structure Global Gene expression: microarray (genechip) data http://www.arabidopsis.org/tools/bulk/microarray/index.html Protein Specific Domain analysis: CDD (Conserved Domain Database) @ NCBI http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml PFAM http://pfam.wustl.edu/ SMART http://smart.embl-heidelberg.de/ Patterns & Profile Post-translational modification, O-glycosylation sites prediction YinOYang http://www.cbs.dtu.dk/services/YinOYang/ NetOGlyc http://www.cbs.dtu.dk/services/NetOGlyc/ Subcellular localization prediction SignalP: Prediction of signal peptide cleavage sites http://www.cbs.dtu.dk/services/SignalP/ ChloroP: Prediction of chloroplast transit peptides http://www.cbs.dtu.dk/services/ChloroP/ MITOPROT: Prediction of mitochondrial targeting sequences http://www.mips.biochem.mpg.de/cgi-bin/proj/medgen/mitofilter TargetP: Prediction of subcellular location http://www.cbs.dtu.dk/services/TargetP/ Transmembrane domains SOSUI http://sosui.proteome.bio.tuat.ac.jp/sosuiframe0.html TMpred http://www.ch.embnet.org/software/TMPRED_form.html Alignment & sequence features of 2 (or more) seq http://us.expasy.org/tools/sim-prot.html (general info: LANVIEW: http://us.expasy.org/tools/lalnview.html) NetPhos - Prediction of Ser, Thr and Tyr phosphorylation sites in eukaryotic proteins Structure: primary, secondary, tertiary (not covered in this lab) Best Plant Biology DBs: (some of these reference other organisms too) TAIR http://www.arabidopsis.org/home.html 7 TIGR http://www.tigr.org/ MIPS http://mips.gsf.de/ Best general DBs: Sequences analysis and related tools Expasy http://www.ncbi.nlm.nih.gov NCBI http://us.expasy.org/ Odd (but still useful) DBs Allergen sequence db Allergome - A db of allergenic molecules Botanical catalogs (from Expasy) NAL - AGIS - Access to many plant genome databases at the National Agricultural Library Mendel - Plant gene nomenclature database from CPGN Arabidopsis Swiss-Prot list - Links to A.thaliana WWW sites and to Swiss-Prot entries TAIR - The Arabidopsis Information Resource MATDB - MIPS Arabidopsis thaliana db TIGR At - TIGR Arabidopsis thaliana db Arabidopsis ABC transporters AMPL - Arabidopsis Membrane Protein Library Arabidopsis at PlaCe - Site with info on P450, glucosyltransferases, etc. Gramene - A comparative mapping resource for grains RGP - Rice Genome Research Program Oryzabase - Japanese rice genome db Rice genome project at Wisconsin Rice-research.org - Monsanto rice genome site TIGR OsGI - TIGR Rice Genome project BeanGenes - Beans genome db BeanRef - Beans genome db and other resources 8 Chlamydomonas resource center ChlamyDB - Chlamydomonas reinhardtii genome db CottonDB - Cotton genome db MaizeDb - Maize genome db INRA Maize - INRA Maize genome db Pisum sativum (pea) web site TIGR Potato - TIGR Potato Functional Genomics project TIGR GmGI - TIGR Soybean Gene Index Snapdragon (A.majus) web site SorghumDB - Sorghum genome db SoyBase - Soybean genome db SoyBase metabolic db - Metabolic subset of the soybean genome db TIGR LeGI - TIGR Tomato Gene Index Dendrome - Forest trees genome db Further reading: Pedestrian guide to analysing sequence databases (oldish, but good general info) http://cubic.bioc.columbia.edu/papers/1999_pedestrian/paper.html Current Protocols in Bioinformatics (subscription required, capmus computers/IP #s should e fine): http://www.mrw2.interscience.wiley.com/cponline/tserver.dll?command=doGetD oc&sUI=&database=CP&useScheme=WIS_Framed.Scheme&getDoc=cp_toc_fs.html