Services Available through the Core (Word)
advertisement
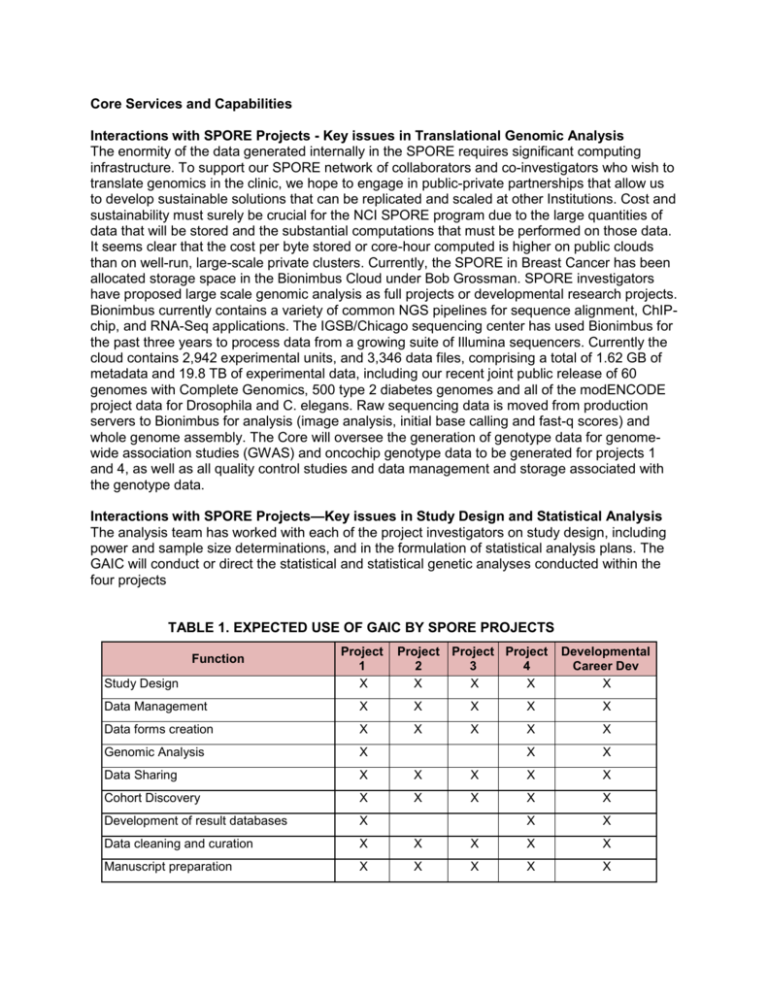
Core Services and Capabilities Interactions with SPORE Projects - Key issues in Translational Genomic Analysis The enormity of the data generated internally in the SPORE requires significant computing infrastructure. To support our SPORE network of collaborators and co-investigators who wish to translate genomics in the clinic, we hope to engage in public-private partnerships that allow us to develop sustainable solutions that can be replicated and scaled at other Institutions. Cost and sustainability must surely be crucial for the NCI SPORE program due to the large quantities of data that will be stored and the substantial computations that must be performed on those data. It seems clear that the cost per byte stored or core-hour computed is higher on public clouds than on well-run, large-scale private clusters. Currently, the SPORE in Breast Cancer has been allocated storage space in the Bionimbus Cloud under Bob Grossman. SPORE investigators have proposed large scale genomic analysis as full projects or developmental research projects. Bionimbus currently contains a variety of common NGS pipelines for sequence alignment, ChIPchip, and RNA-Seq applications. The IGSB/Chicago sequencing center has used Bionimbus for the past three years to process data from a growing suite of Illumina sequencers. Currently the cloud contains 2,942 experimental units, and 3,346 data files, comprising a total of 1.62 GB of metadata and 19.8 TB of experimental data, including our recent joint public release of 60 genomes with Complete Genomics, 500 type 2 diabetes genomes and all of the modENCODE project data for Drosophila and C. elegans. Raw sequencing data is moved from production servers to Bionimbus for analysis (image analysis, initial base calling and fast-q scores) and whole genome assembly. The Core will oversee the generation of genotype data for genomewide association studies (GWAS) and oncochip genotype data to be generated for projects 1 and 4, as well as all quality control studies and data management and storage associated with the genotype data. Interactions with SPORE Projects—Key issues in Study Design and Statistical Analysis The analysis team has worked with each of the project investigators on study design, including power and sample size determinations, and in the formulation of statistical analysis plans. The GAIC will conduct or direct the statistical and statistical genetic analyses conducted within the four projects TABLE 1. EXPECTED USE OF GAIC BY SPORE PROJECTS Function Study Design Project 1 X Project Project Project 2 3 4 X X X Developmental Career Dev X Data Management X X X X X Data forms creation X X X X X Genomic Analysis X X X Data Sharing X X X X X Cohort Discovery X X X X X Development of result databases X X X Data cleaning and curation X X X X X Manuscript preparation X X X X X