Chloroplast Experimental Paper
advertisement

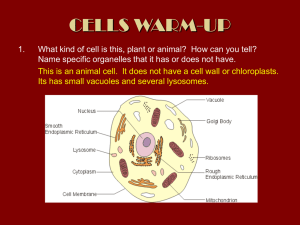
Elkhatib 1 Wiaam Elkhatib Britt Reese, Honors Biology Lab K325 04/30/2014 Chloroplast Research Paper ABSTRACT The study of protein localization in chloroplasts can give insight on the workings of photosynthesis and potentially serve as disease models for other organisms. Chloroplasts were homogenized and underwent centrifugation to fractionate broken, whole, stromal, and thylakoid chloroplast portions with the goal of determining protein localization based on molecular weights. Fractions were analyzed using an SDS page to determine which proteins were expressed in specific localizations. Intact chloroplasts were found to have well distributed bands on an SDS page, while broke chloroplasts were spread out to the extremes. Various proteins were found to be present in both thylakoids and stromal extract fractions, while others were found to be in the broken chloroplast fractions. Further testing and review will be required to affirm the specific findings. Results can later be analyzed via alternative processes to help gain insight on protein roles during plant photosynthetic pathways. INTRODUCTION Photosynthesis is a process used by plants and some other organisms to convert energy from the sun in the form of light into chemical energy that can be processed and used to fuel organismal processes (Lodish). This chemical energy is stored as carbohydrates, such as sugars, which are synthesized from carbon dioxide and water molecules. Oxygen is generally released into the Elkhatib 2 environment as a waste product. The overall purpose of photosynthesis is to maintain atmospheric oxygen levels and supply energy to higher level eukaryotes, whereas moss, as well as spinach leaves, can be used as subjects for chloroplast isolation (Lang). Two different types of reactions partake in photosynthesis, known as light and dark reactions. In light reactions, which occur in thylakoids, a photon of light strikes an electron in a pigment molecule, and the energy is passed between molecules thereafter. The reaction center is reached, which has a special form of chlorophyll (P700). The chlorophyll at the reaction-center of photosystem II is called P680, where the absorbed light energy powers an oxidation-reduction reaction. Light thus drives the synthesis of NADPH2 and ATP, the two main products of the light- dependent reactions, by energizing these photosystems. Lastly, the removal of hydrogen and electrons from water by photosystem II in the light produces O2, the final product of the light reactions of photosynthesis (Lodish). In dark reactions, carbon dioxide and other compounds are converted into glucose. These reactions occur in the stroma, outside of the thylakoid membranes, and are a continuation of the light reactions previously described. Three phases to the dark reactions, which in order, are the Calvin cycle, reduction reactions, and RuBisCo regeneration. In the first stage of the Calvin cycle, CO2 molecules come into play, where they each use two molecules of ATP and two molecules of NADPH (produced earlier in the light-dependent stage). First, ribulose bisphosphate binds CO2 from the stroma in a process called CO2 fixation, which is catalyzed by RuBisCo. The molecule synthesized degrades into two molecules of the three-carbon 3phosphoglycerate (3PG). Next, the two 3PG molecules are converted into glyceraldehyde 3phosphate (G3P) molecules using ATP, then separating the phosphate bond and adding the hydrogen gotten from NADH2. Lastly, a total of six molecules of G3P are ultimately made. Only Elkhatib 3 one of these molecules exits the cycle while the rest enter a complicated process that regenerates RuBisCo, and two molecules of G3P later combine to form one molecule of glucose as the cycle continues and repeats. Different proteins also aid in these processes and are elaborated on below as discussed by their structural localizations. Chloroplasts have a fairly interesting and complex structure which consists mainly of thylakoids, stroma, a double membrane, granum, lamella, and the lumen. These structures have multiple processes associated with photosynthesis and generation of fuel for the cell which occur within them, most of which have been described above, and allow the chloroplast to be importcompetent (Aronsson). Figure I1 depicts the structure of a chloroplast with associated major structures labeled. Elkhatib 4 Each structure also is associated with certain proteins which are later tested in this laboratory exercise. While a plethora of proteins can be found in chloroplasts, only the major ones will be discussed. NADPH oxidoreductase(FNR) is a binding enzyme encoded by a small nuclear gene family in higher plants, and transfers an electron from each of two ferredoxin molecules to a single molecule of NADPH found in photosystem 1 during light reactions in both thylakoids and stroma. “The binding of the FNR to the integral membrane proteins on the thylakoid membrane is enhanced under acidic conditions, so recruitment and binding of FNR to the thylakoid membrane may be a method of storing and stabilizing the enzyme in the dark when photosynthesis is not occurring (Benz).” In addition to activating enzymes of the Calvin cycle, the thioredoxin activates enzymes of other chloroplast processes such as the C4 cycle, photophosphorylation, and mRNA translation. HP15 is a putative membrane channel protein, likely to be located in the outer membrane of the chloroplast, as are other “HP” type proteins. VATA, HP54, and PSAL all are involved in the electron transport chain of thylakoids. Stromal proteins which aid in translation include RR18 ribosomal protein and EFTU elongation factor. Some proteins still have unclassified functions within the chloroplast, such as HP45 and RCP1 (Ferro). Some of the few known transporters of the chloroplast envelope (IEP30, IEP45, IEP33, IEP62, and IEP60) are already known to be located in the inner membrane. Many others participate in dark reactions of the stroma and light reactions of thylakoid, or assist in transport and translation/transcription in the stroma. To analyze these components and proteins, centrifugation and SDS PAGE processes were done. Differential centrifugation is a common procedure in biology that is used to separate organelles from whole cells in order to better study them. It involves tissue sample being first homogenized to break the cell membranes and then mix up any contents within the cell (Bio-Rad). The Elkhatib 5 homogenate is centrifuged many times, giving a supernatant (liquid part) and solid (pellet). The theory is that heavier, larger compounds will clump and pellet at the bottom while lighter, smaller compounds will remain in the supernatant, a process known more specifically as density separation. The proteins or cellular particles to be studied can then undergo SDS page analysis (Rabilloud). This procedure involves using buffers and other chemicals in order to denature proteins and place them in SDS (also a denaturant) in separate channels. The goal of this step is to ensure than proteins are unfolded and thus only travel through the medium based on charge and length/weight. An electric current is then run through the gel, causing particles to move further if smaller and less far if heavier. In separating them by molecular weight, proteins can be identified by comparison on standard curve of the log of the weight verses the Rf (retention factor), which is a unit-less ratio of distance traveled over total distance. Relative distances traveled are theoretically only affected by the length, charge, or conformation of the molecule at hand. The goal of this experiment is to identify major chloroplast proteins by determining the relative molecular weights. The hypothesis is if chloroplasts are lysed and run on an SDS page, the whole chloroplasts and thylakoids will have lower mobility than the broken chloroplasts and stromal extracts. Consequently, it can be inferred that the proteins which have higher molecular weights would move less along the SDS page, while those with lower molecular weights will move further along. Whole and broken chloroplasts should both have wide ranges of motility due to having many different components, but the broken chloroplasts should have clearly visible bands at the upper and lower extremes of motilities (since they will break into a group of small and large fractions) while the whole will exhibit gradual, distinct bands uniformly, similar to the ladder. There is a correlation between the molecular weight of the proteins and their location Elkhatib 6 within the chloroplast as well, involving the proteins discussed above and outlined further by Ferro’s Molecular and Cellular Proteomics research paper (Ferro). Those proteins associated with transport can be found in all chloroplast portions, and proteins associated with the photosystems and transcription/translation should be found in their respective portions (ex. Dark reaction-related proteins exist in stroma, etc.). Naturally, there may be some overlap between the proteins and their respective organelle location due to their having various different functions or perhaps contamination. MATERIALS AND METHODS The following list describes the array of materials used in the methodology described below. All materials and methods were designed and followed by information previously provided by a polyacrylamide gel and electrophoresis detection guide (Bio-Rad). Materials included: 250-mL polypropylene centrifuge bottle Beckman JA-14 rotor 50-mL centrifuge tubes Parafilm Distilled water 15-mL Corex tube Acetone 50-mL polycarbonate centrifuge tube Percoll Gradient Solution Beckman JA-20 rotor Elkhatib 7 Spinach leaves Cold deionized water 600-mL beaker Ice cold Grinding Buffer Pre-chilled Waring blendor and jar Cheesecloth Funnel 12x75 mm glass tubes Genesys 5 Chloroplast Lysis Buffer 1.5-mL microfuge tubes SDS-PAGE Sample Buffer BioRad Mini-Protean II gel electrophoresis device 95°C water bath 12.5% polyacrylamide gel Coomassie Blue Staining Solution Destaining Solution Purification and Fractionation of Spinach Chloroplasts Approximately 20g of spinach leaves were washed with cold, deionized water and cut up into roughly 1 cm x 1 cm squares. The obtained leaf material was weighed (20.02g) and suspended in 200 mL of ice cold Grinding Buffer in a beaker. This was then transferred to a pre-chilled Elkhatib 8 Waring blender and homogenized until no visible leaves were present in the jar. The green homogenate was then strained through four layers of cheesecloth into a 250-mL polypropylene centrifuge bottle and the filtrate centrifuged at 4,000 rpm in a Beckman JA-14 rotor for 5 minutes at 4°C. After discarding the supernatant, 8 mL of the Grinding Buffer was added to the chloroplast pellet for re-suspension. Next, 8 mL of the chloroplast suspension was layered on top of the Percoll gradient and centrifuged at 8,500 rpm in a Beckman JA-20 rotor for 15 minutes at 4°C. After centrifugation, the gradient displayed an upper band containing broken chloroplasts and a lower band containing intact chloroplasts, which were separated into 50-mL centrifuge tubes. Grinding Buffer was added to each tube to remove remaining Percoll until their weights were nearly equal. The contents were mixed and centrifuged in the JA-20 rotor for 4 minutes at 4°C. Supernatants were again discarded and pellets re-suspended in Grinding Buffer. 500 µL of distilled water and 4 mL of acetone were added to 500 µL of each suspension in 15-mL Corex tubes, which then underwent vortexes, kept on ice, and centrifuged at 9,000 rpm in the JA-20 rotor for 5 minutes. 1 mL of each supernatant was transferred to a tube to undergo absorbance readings against an 80% acetone blank using a Genesys 5. The amount of chlorophyll was calculated using the following equation: Chl (in µg/mL) = [20.2(A645) + 8.02(A663)] x dilution factor [Equation 1] The remaining fractions of intact and broken chloroplasts were re-pelleted by centrifuging at 4,500 rpm in the JA-20 rotor for 4 minutes at 4°C. The pellets were re-suspended in Chloroplast Lysis Buffer and incubated on ice for 15 minutes with interruptions to vortex the suspensions every 5 minutes. 200 µL aliquots of the lysed whole and broken chloroplasts were separated and kept in 1.5-mL microfuge tubes for later use. From the intact chloroplasts, two more 200 µL Elkhatib 9 aliquots were collected and centrifuged. The supernatants containing the stromal extracts were collected and the pellets were given 200 µL of Chloroplast Lysis Buffer, re-suspended, and recentrifuged. The process was repeated and the resulting re-suspended pellets contained thylakoid membranes. At the end of this portion of the procedure, there were two 200 µL aliquots each of whole chloroplasts, broken chloroplasts, stromal extracts, and thylakoid membranes. Recovery of Chloroplast Proteins To recover chloroplasts, 800 µL of ice cold acetone was added to each of the above samples for acetone precipitation. The contents of the tubes were mixed and kept on ice for 20 minutes, following which they were centrifuged at top speed for 5 minutes. The supernatants were discarded and 1 mL of ice-cold 80% acetone was added to the tubes before re-centrifugation. The tubes were drained of supernatant through evaporation and 200 µL of SDS-PAGE Sample Buffer was added to each formed pellet to be re-suspended. SDS-PAGE of Chloroplast Proteins The proteins were denatured in a 95°C water bath in preparation for SDS-PAGE using a Bio-Rad Mini-Protean II gel electrophoresis machine. 20 µL of each sample was loaded onto a 12.5% polyacrylamide gel and allowed to run for half an hour. Lastly, the gel was placed in Coomassie Blue Staining Solution for 30 minutes, followed by for 1-3 hours and eventual drying of the gel to be later analyzed. Analysis of the Stained Cells The mobility of the bands formed on the gel was measured and the graphical relationship between the log of the molecular weight and the relative mobility of the proteins was plotted to Elkhatib 10 calculate the molecular weights from the curve. The bands were compared to find which proteins were displayed in each chloroplast fraction. Figure M1 above depicts a flowchart of the overall chloroplast protein isolation procedure, starting from leaves and ending with respective SDS page samples. RESULTS Elkhatib 11 Following SDS PAGE analysis and comparison of bands to standard curve graphs and previously determined ladder band values, the results were compiled. The resulting SDS PAGE highlighted important band mobility values and is shown in Figure R1, with further elaboration in Table R1. Figure R1 above shows the SDS PAGE results, with the right two bands being the ladders. Each column is labeled as which fraction it corresponded to in Table R1. Row # 1 2 3 4 5 6 7 L1 L2 Component Whole Whole Thylakoid Stroma Stroma Stroma Broken Ladder Ladder Table R1: The SDS-PAGE column data is outlined for Figure R1, with numbers labeled one through seven (as well as ladders) going from left to right in the figure above. Elkhatib 12 The band values were then compared to pre-determined values taken from the manual (Bio-Rad) and previous research publications. This is shown in Figure R2-1 and the references section. Figure R2-1 depicts the comparative ladder used to determine the standard curve. In order to analyze these fractions of the spinach chloroplasts, a standard curve was generated using molecular weights of the above in Figure R2-1proteins used and their corresponding retention factors (Ferro). The corresponding found equation was: 𝑦 = −1.638𝑥 + 5.4598 [𝐄𝐪𝐮𝐚𝐭𝐢𝐨𝐧 𝟐] Elkhatib 13 log(MW) VS Retention Factor 7 log (molecular weight) 6 5 4 y = -1.638x + 5.4598 R² = 0.7766 3 2 1 0 0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1 Retention Factor Figure R3: A standard curve depicting log of molecular weight against retention factor for chloroplast protein analysis and corresponding Equation 2. Finally, the molecular weights and retention factors of the standard curves were compared to known values associated with previously tested proteins in the literature (see references). The below table is separated by row. Molecular weight was found by plugging in respective retention factor values as the “y” variable of Equation 2 and solving for the log molecular weight, the “x” variable. Inverse log of this “x” value yielded molecular weight in units of Daltons. Known molecular protein weights were matched with the calculated weights. Major band identities are also shown. Not all molecular weights matched exactly with known values. Row 1 (Whole) RF Value 0.217 0.3 0.43 log(MW) 5.104354 4.9684 4.75546 MW 127161.0192 92982.23906 56945.57729 Protein? HP112 OEP86 HP56b Elkhatib 14 0.5 0.717 0.8 0.95 4.6408 4.285354 4.1494 3.9037 43732.06652 19290.96708 14105.87399 8011.24475 PORB NADPH oxidoreductase THM1 thioredoxin AP15 membrane channel OEP6 Row 2 (Whole) RF Value 0.217 0.3 0.417 0.5 0.63 0.7 0.783 0.97 log(MW) 5.104354 4.9684 4.776754 4.6408 4.42786 4.3132 4.177246 3.87094 MW 127161.0192 92982.23906 59807.27292 43732.06652 26783.04802 20568.37587 15039.93641 7429.164931 Protein? HP112 OEP86 TIC55 PORB NADPH oxidoreductase HP25 RB52 HP15 OEP6 Row 3 (Thylakoid) RF Value 0.217 0.3 0.45 0.5 log(MW) 5.104354 4.9684 4.7227 4.6408 MW 127161.0192 92982.23906 52808.03408 43732.06652 Protein? HP112 OEP86 HP53 PORB NADPH oxidoreductase Row 4 (Stroma) RF Value 0.23 0.3 0.45 0.5 log(MW) 5.08306 4.9684 4.7227 4.6408 MW 121076.5395 92982.23906 52808.03408 43732.06652 Protein? Unknown 1 OEP86 HP53 PORB NADPH oxidoreductase Row 5 (Stroma) RF Value 0.23 0.3 0.45 0.5 log(MW) 5.08306 4.9684 4.7227 4.6408 MW 121076.5395 92982.23906 52808.03408 43732.06652 Protein? Unknown 1 OEP86 HP53 PORB NADPH oxidoreductase Row 6 (Stroma) RF Value 0.23 0.317 log(MW) 5.08306 4.940554 MW 121076.5395 87207.5328 Protein? Unknown 1 HP87 Row 7 (Broken) RF Value log(MW) MW Protein? Elkhatib 15 0.13 0.167 0.23 0.53 0.9 1 5.24686 5.186254 5.08306 4.59166 3.9856 3.8218 176546.8609 153551.4776 121076.5395 39053.50344 9673.8645 6634.374758 PSAG HP15 Unknown 1 HP40b Unknown 2 OEP6 L1 & L2 (Ladders) RF Value 0.066666667 0.133333333 0.15 0.216666667 0.366666667 0.516666667 0.716666667 0.883333333 0.933333333 log(MW) 5.301029996 5.065392962 4.988558957 5.820857989 4.653212514 4.491361694 4.322219295 4.158362492 3.812913357 MW 200000 116250 97400 66200 45000 31000 21000 14400 6500 Protein Unknown 3 HP112 OEP86 HP65b HP45b HP30c RBS2 HP15 Unknown 4 Table R2: This table is indicative of various tested proteins with their respective band motilities and descriptors of major bands. Molecular weights are shown in Daltons. The amount of chlorophyll in each band was also calculated using Equation 1. For the upper band, the A645 absorbance value was found to be 0.050 and the A663 value was 0.046, all with a dilution factor of 10, giving a chloroplast concentration of 13.79µg/ml. For the lower band, the same respective absorbance values were found to be 0.079 and 0.075, also having a dilution factor of 10, yielding a chloroplast concentration of 21.97µg/ml. Certain inconsistencies and assumptions were made in the pairing of band value to protein identity, due to potential fallacies in the carrying out of the procedure which requires further discussion. DISCUSSION Elkhatib 16 The goal of this experiment was to identify major chloroplast proteins by determining the relative molecular weights, as depicted in Table R2 and Figure R3. The hypothesis previously stated was, for the most part, validated by the results procured. However, some inconsistencies and details deferred from expected results or yielded little information to support the hypothesis as a whole. When chloroplasts were lysed and run on an SDS page, the whole chloroplasts and thylakoids were shown to have lower mobility than the broken chloroplasts and stromal extracts. Consequently, the SDS page affirmed that the proteins which have higher molecular weights would move less along the SDS page, while those with lower molecular weights will move further along. Whole and broken chloroplasts both indeed did have wide ranges of motility due to having many different components, while the broken chloroplasts had clearly visible bands at the upper and lower extremes of motilities (since they broke into a group of small and large fractions) . Higher chloroplast concentration was found in the whole chloroplast region than the broken. The whole chloroplasts exhibited gradual, distinct bands uniformly, similar to the ladder, as can be seen by the values depicted in Table 2. Naturally, there was some overlap between the proteins and their respective organelle locations though. Intact chloroplasts were found to have well distributed bands on an SDS page, while broken chloroplasts were spread out to the extremes. Some molecular weights had larger or smaller than expected ranges. Thylakoid bands also matched up with stromal, suggesting that they have similar molecular weights, but on the contrary, it is already understood that thylakoids are much heavier than stroma since they pelleted first during centrifugation and the stroma was left in the supernatant. Previous research on the Proteomics of the Chloroplast Envelope Membranes from Arabidopsis thaliana affirms the following findings but does not exactly match up with the Elkhatib 17 molecular weights (Ferro). Most localization was found to be accurate, according to previous literature. For most cases, the previously established molecular weights of the proteins in literature corresponded to the weights found using the standard curve (see Table R2). As seen by Table R2, proteins predicted to be located in the thylakoid are HP112 (protein import), OEP86 (chloroplast outer envelope import protein), HP53 (unknown function), and PORB NADPH oxidoreductase (assist in light reactions). NADPH oxidoreductase is necessary for electron transport in light reactions, where light reactions occur in thylakoids, so it makes sense for it to be found there. Molecular chaperones and import proteins are also present in the thylakoid lumen, thus justifying the presence of such proteins found here. HP53 has a function that is not yet known, but it is likely involved in protein transport or photosystems. Proteins identified to be in the stroma are Unknown 1, OEP86, HP53, PORB NADPH oxidoreductase, and HP87. NADPH oxidoreductase is necessary for electron transport in light reactions, but dark reactions occur in the stroma. However, “the binding of the FNR to the integral membrane proteins on the thylakoid membrane is enhanced under acidic conditions, so recruitment and binding of FNR to the thylakoid membrane may be a method of storing and stabilizing the enzyme in the dark when photosynthesis is not occurring (Benz).” The disruption of this binding via the procedure could have caused FNR to be unbound and found in the stroma as well. Because OEP86 functions as an import receptor of the chloroplast’s outer envelope, it may have been mixed in with the stroma due to the closeness of the chloroplast’s membranes and the stroma itself. Similar reasoning justifies the locations of HP53 and HP87 proteases and chaperones. Unknown 1 may have import functionality or assist in photosynthesis or translation/transcription (which occurs in the stroma). Elkhatib 18 Whole and broken chloroplasts should have many similar proteins found elsewhere in the stroma and thylakoids, but with heavier bands and perhaps more import proteins found on the outer membrane. The whole chloroplast portion was found to have HP112, OEP86, HP56b, PORB NADPH oxidoreductase, THM1 thioredoxin, AP15 membrane channel, and OEP6. The membrane channel with HP112 and HP56b are different types of import proteins which were expected to be found in the whole chloroplast portion. OEP86 chloroplast outer envelope import protein falls into that same category. NADPH oxidoreductase is necessary for electron transport in light reactions, which occur in both thylakoids and could have mixed into the stroma, so it makes sense for it to be found in the whole chloroplast too. “Thioredoxin effectively activates NADP–malate dehydrogenase, an enzyme of the C4 photosynthetic carbon cycle and C3 redox shuttle, and deactivates glucose-6-phosphate dehydrogenase. Plants have a third class of thioredoxin, the h type—located in the cytosol, endoplasmic reticulum, and mitochondrion that is reduced by NADPH in a reaction catalyzed by a flavoprotein enzyme, NADP–thioredoxin reductase (Lodish).” Surely this protein is present in various locations in the chloroplast, and unsurprisingly is identified. The broken chloroplast portion also contained some transport and import proteins, along with a few unknown. The exception was with PSAG, also known as photosystem I reaction center subunit V. This should have been found in the thylakoid membrane, where photosystem I and II take place, but may have been lost or exhibited very lightly due to there being a very low concentration of it during the procedure. The ladder displayed an array of proteins which were found in previous chloroplast fractions, with some unmatched which simply could not be identified. Examples would be RBS2, along with the unknowns 3 and 4. RBS2 is known to be a ribosome binding site (Lodish), which also plays a factor in the stroma via translation. Elkhatib 19 CONCLUSION Intact chloroplasts were found to have well distributed bands on an SDS page, while broken chloroplasts were spread out to the extremes. Overall, the results predicted proteins to be located in the thylakoid as HP112 (protein import), OEP86 (chloroplast outer envelope import protein), HP53 (unknown function), and PORB NADPH oxidoreductase (assist in light and dark reactions). Proteins identified to be in the stroma are Unknown 1, OEP86, HP53, PORB NADPH oxidoreductase, and HP87. Each protein had its individual function. Because some of the molecular weights only approximately lined up with previously determined values, it became apparent that some key sources of error played a role in the outcome. The major source of error in this experiment occurred during the procedure. Chloroplast fractions were supposed to have been heated at 95ºC for 4 minutes, but instead had their temperature set much higher and the tubes holding the solutions had their lids blown off well before the 4 minute mark. This means that not only were the solutions potentially not activated or neutralized as necessary, but that the proteins were not denatured to the extent required. It is believed that because thylakoids are much larger (and thus have more bonds) than stroma, they denature much less quickly and ended up having a lower surface area than stroma at the time of removal. This would have caused the thylakoid fractions to travel further than they should have on the SDS page (since their lower than usual surface area meant less friction in the gel medium), causing the illusion that thylakoids were just as heavy as stroma. One other potential error source was contamination of the SDS page channels during injection of the protein solutions. Loading excess solution or having shaky hands can cause some solution to spill out and mix with others, perhaps causing proteins that should have been in stromal extract to end up in other chloroplast fractions or visa-versa. Mislabeling might also have played a part, because the thylakoid and Elkhatib 20 stromal fractions seemed identical, it is possible that the previously discussed misplaced stromal fraction may have indeed been the thylakoid fraction, and thus meaning that there was no thylakoid fraction analyzed in reality. Looking at the standard curve, an R2 value of 0.7766 means that the log of molecular weight vs. retention factor did not correlate perfectly in a linear fashion as necessary and opens the possibility of flawed data in the form of too broad of range of molecular weight. This would cause some errors in the protein identifications. Future-wise, the molecular weights of the proteins tested would be compared with even more recent sources, chloroplast fractions would be denatured at the correct temperature and time length, and more care would be taken to prevent possible contamination or mislabeling between fractions. Other methods can be used to further characterize the band identities as well. Spectroscopy could be run on the solutions and analyzed to determine the exact components present in each fraction. A more in-depth literature review could also be conducted to research all past experiments published on the specific chloroplast proteins tested and compared to the results. To officially confirm the identity of the bands, however, the best option would be to send the SDS page and solutions back to the company of purchase and request that they undergo a bioinformatics analysis to determine their genetic code sequence. That would affirm exactly what proteins were truly in the solutions and whether or not any contamination of unknown factors came into effect. REFERENCES Aronsson, Henrik. “A Simple Method for Isolating Import-competent Arabidopsis Chloroplasts.” University of Leicester. 17 September, 2002. Web. 14 April 2014. Elkhatib 21 Benz, JP.; Lintala, M.; Soll, J.; Mulo, P.; Bölter, B. (Nov 2010). "A new concept for ferredoxinNADP(H) oxidoreductase binding to plant thylakoids". Trends Plant Sci 15 (11): 608–13. Bio-Rad. “A Guide to Polyacrylamide Gel Electrophoresis and Detection.” Bio-Rad Laboratories, Inc. Print. 15 April, 2014. Ferro, Myriam. “Proteomics of the Chloroplast Envelope Membranes from Arabidopsis thaliana.” Molecular and Cellular Proteomics. 28 May, 2003. Print. 14 April, 2014. Lang, Erika. “Simultaneous isolation of pure and intact chloroplasts and mitochondria from moss as the basis for sub-cellular proteomics.” Springer. 20 October 2010. Web. 14 April, 2014. Lodish, Harvey. “Molecular Cell Biology.” W.H. Freeman. 1986. Print. 14 April, 2014. Rabilloud, Thierry. “Variations on a Theme: Changes to Electrophoretic Separations that can Make a Difference.” Biochemistry and Biophysics of Integrated Systems, CEA Grenoble. 2011. Web. 14 April, 2014.