tpj12913-sup-0020-AppendixS7
advertisement

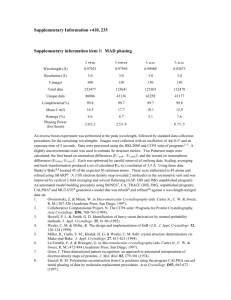
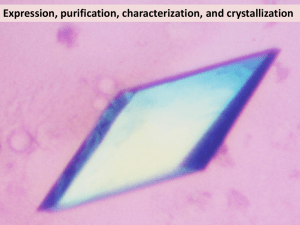
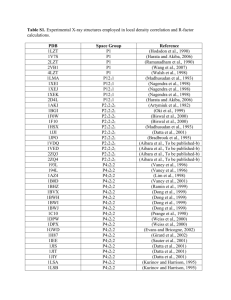
Methods S7: Crystallization and resolution of the structure Crystals for the native and selenomethionine labeled proteins were obtained at 19°C from a 1:1 ml mixture of a 7 mg/ml protein solution (stored in 50mM sodium phosphate pH 7, 300mM NaCl) with crystallization solutions composed of 1.2M Na/K tartrate, 100 mM Tris pH8 or 1.6M ammonium sulfate, 0.1M Hepes pH7, respectively. For data collection, the crystals were directly flash-cooled in liquid nitrogen. The diffraction data were recorded on beam line Proxima-1 (synchrotron SOLEIL, France). The structure was determined by the multiple wavelength anomalous diffusion (MAD) method using the anomalous signal of the selenium atoms. Data were processed using the XDS package (Kabsch, 1993). The space group was P43212 with two AvrLm4-7 molecules per asymmetric unit. The 4 Se sites (two per monomer) were located using the program SHELXD (Schneider and Sheldrick, 2002). The refinement of these Se sites as well as phasing was performed using the SHARP program. The resulting map was further improved by NCS averaging using the RESOLVE program (Terwilliger, 1999). The model was build semi-automatically using the BUCCANEER software (Cowtan, 2006) and manually using the COOT program (Emsley and Cowtan, 2004). The model was further improved by iterative cycles of manual rebuilding using COOT and refinement using the PHENIX program (Adams et al., 2002). During refinement using data processed in space group P43212, the Rfree factor did not drop below 35% despite electron density maps of excellent quality. The analysis of the crystal packing revealed the presence of a crystallographic homotetramer formed between the two molecules of the asymmetric unit and a neighboring one via a crystallographic two-fold axis. To test whether the high value of Rfree factor could be due to the presence of pseudo symmetry, we processed the diffraction data in space group P43 with four molecules per asymmetric unit as described above. The four molecules of the asymmetric unit were located by molecular replacement using the MOLREP program (Vagin and Teplyakov, 1997) and this model was refined using PHENIX. This yielded a Rfree factor of 30% without any addition of water molecules or model improvement for both datasets. Following this observation, the final model was refined using the data processed in space group P43. The final model contains residues 5 to 134 from chains A and C as well as residues 7 to 133 from chains B and D. In the structure refined against the dataset collected from crystals grown in tartrate as precipitant, 180 water molecules and four tartrate molecules have been modeled. In the structure refined against the dataset collected from crystals grown in ammonium sulfate as precipitant, 260 water molecules have been modelled. Statistics for data collection and refinement are summarized in Table 1. The atomic coordinates (and structure factors) have been deposited into the Brookhaven Protein Data Bank under the accession numbers 4FPR and 4FPQ. Adams, P.D., Grosse-Kunstleve, R.W., Hung, L.W., Ioerger, T.R., McCoy, A.J., Moriarty, N.W., Read, R.J., Sacchettini, J.C., Sauter, N.K. and Terwilliger, T.C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr., 58, 1948-1954. Cowtan, K. (2006) The Buccaneer software for automated model building. Acta Crystallogr. D Biol. Crystallogr., 62, 1002-1011. Emsley, P. and Cowtan, K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr., 60, 2126-2132. Kabsch, W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr., 26, 795-800. Schneider, T.R. and Sheldrick, G.M. (2002) Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr., 58, 1772-1779. Terwilliger, T.C. (1999) Reciprocal-space solvent flattening. Acta Crystallogr. D Biol. Crystallogr., 55, 1863-1871. Vagin, A. and Teplyakov, A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Cryst., 30, 1022-1025.