Project Summary_Rao_..
advertisement

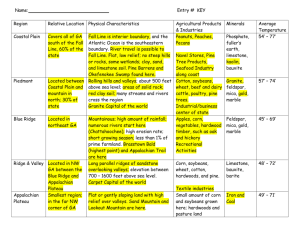
Arjun Rao Dat Tran Patrick Duffy STATS 512 Project Proposal Understory Diversity in Pine and Hardwood Forested Sites as Compared to Overstory Area, pH, Cation Concentration, and Litter Depth. Project Summary: Pine forest sites and hardwood forest sites have different ground-layer diversity (in # of plants and proportion of each species). This will be due to a) difference in basal area of overstory trees b) deeper litter depth in pine plantations, or c) lower nutrient quality in pine plantations, any of which may lead to lower diversity in pine plantations. Some Background Pine litter decomposes more slowly due to high concentrations of tough materials. This means that pine plantations have large amounts of pine litter. This may stop tree seedlings from establishing because of inaccessibility of light at the mineral soil surface or inability of the seed to reach the mineral surface. When decomposing, the needles release H+, which leads to lower pH, as well as release and dissolve cations (nutrients for trees). These nutrients can be leeched (removed) from the surface soil and become unavailable to plants, causing lower plant abundance. It can also cause sandier soil over time. Pine plantations may shade more light out if their basal area is higher, and if their leaf area is less. This is because larger trees intercept more light and therefore leave less to the understory. This will be measured indirectly (as a simplification) using basal area. Analysis The response variable will be understory diversity, and the explanatory variables will be Overstory Basal Area, litter depth, pH, and cation concentration. We will calculate species diversity as the Shannon-Weiner index (H’). We will then use the Kolmorogov-Smirnov test for goodness of fit to check for normality of each dataset. Assuming the data are not normal, we will use non-parametric tests such as KurskalWallis ANOVA on ranks with Dunn’s pairwise multiple comparison test (Jenkins and Parker, 1999). We will run t tests with each explanatory variable alone with the response variable to check for significance. If a variable does not show a significant linear relationship with diversity, it will be discarded. Then, an ANOVA will be performed to check for the similarity of these variables, to aid in establishing which of the variables are independently affecting diversity and which may be working concurrently. Model Selection 1. Run simple linear regression on explanatory variables (Basal Area, Litter depth, pH, cation) to response variable (H’), checking for linear relationship. 2. Diagnose with t test p values, QQ plots, X vs Y, X vs Residual, and residuals vs time plots. Checking for normality, linear relationship, constant variance of error terms, and independence of error terms. 3. Remove or transform variables that violate one or more of the above assumptions. 4. Run ANOVA with all variables. Check for difference in explanatory variables to each other as well to ensure that each variable determines something meaningful with respect to diversity (H’). 5. Find final regression model that best explains changes in diversity. Literature Cited Jenkins, Michael A., and George R. Parker. "Composition and diversity of ground-layer vegetation in silvicultural openings of southern Indiana forests." The American midland naturalist 142.1 (1999): 1-16. Jenkins, Michael A., and George R. Parker. "The response of herbaceous-layer vegetation to anthropogenic disturbance in intermittent stream bottomland forests of southern Indiana, USA." Plant Ecology 151.2 (2000): 223-237. Jenkins, Michael A., and George R. Parker. "Woody species composition of disturbed forests in intermittent stream bottomlands of southern Indiana." Journal of the Torrey Botanical Society (2001): 165175.