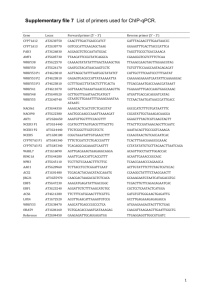
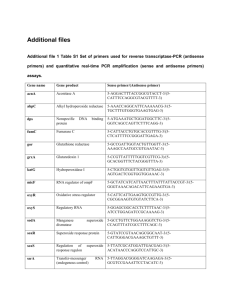
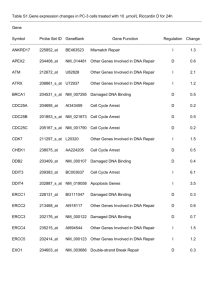
fyr12000-sup-0004-TableS2
advertisement

SUPPLEMENTAL TABLE 2 Primers used in this study Primer OD16 OD21 O276 O277 O278 O279 O280 O245 O246 OD18 OD21 OD51 O780 OD25 OD30 OD52 O780 O274 O290 O294 O295 O588 O589 O272 O291 O304 O343 O580 Sequence Purpose Primers to delete genes and check for deletion tgactgggattccgtgagacggac Delete C. glabrata PHO4 with KANMX6 atcttgtgccctccaaacagaactg aagacccacaaaagaagaataagccaaaacaaaggaaataATGTCT AAAGGTGAAGAATT gtaggccctatctcttcttcaTCGATGAATTCGAGCTCGTT AACGAGCTCGAATTCATCGAtgaagaagagatagggcc tac gtcgtgtcgtgatagaatactg gctaagactacaatgtgcgg GGGACAACACCAGTGAATAATTCTTCACC ATGGAACTGGCAATTTACCA ctgttagtgatgggtgaacaggtcggatccccgggttaattaa atcttgtgccctccaaacagaactg TACGAGTGTAATAATCGTTGG TTAATTAACCCGGGGATCCG gctcgctctccattgcctattgtag tcccatccaatcttatccggtgc TGTCCCGGTACAGCTGCTGC TTAATTAACCCGGGGATCCG ataaaaaccggcacgcaatagtacttattttctgttagtgcacaggaaacagc tatgacc agtagtcatttgaagttttgtcttggagtatctgtagtcaGTTGTAAAA CGACGGCCAGT ataaaaaccggcacgcaatagtacttattttctgttagtgatgggccgtacaa cttctga agtagtcatttgaagttttgtcttggagtatctgtagtcacgtgctcacgttctg ctgta cgatgaccaccacactgttgaggactgtgggattgcgctgcggatccccg ggttaattaa cgttggtgccccgcagacagtccacatgcatggtgatacgGAATTCG AGCTCGTTTAAAC gagatgagcaaaggagacagaacaagagtagcagaaagtccacaggaa acagctatgacc ccgatatgcccggaacgtgcttcccattggtgcacggtcaGTTGTAA AACGACGGCCAGT gagatgagcaaaggagacagaacaagagtagcagaa agtcATGggtgaacaggtagagga ccgatatgcccggaacgtgcttcccattggtgcacggtcatttcttgtactg ctcaagct tgtccagtgcctcatatttacaaagagccgaggctcaccgctctagaact agtggatcc Fusion primers: Delete CgPHO84 with XFP-KANMX6 Fusion primers: Homology to XFP 3' end to delete CgPHO84 with XFPKANMX6 Check primer for CgPHO84 deletion 5' Check primer for XFP - 3' Check primer for XFP - 3' Delete CgPHO4 with NATMX6 Check primer for CgPHO4 deletion PringleChk Delete CgPHO2 with NATMX6 Check primer for CgPHO2 deletion PringleChk Precisely delete CgPHO4 with CgURA3 Replace CgURA3 with ScPHO4 in DG37 Delete CgHIS3 with NATMX6 in DG38 Precisely delete ScPHO4 with ScURA3 Replace ScURA3 with CgPHO4 in DC33 Amplify C. glabrata promoter-YFP constructs to integrate into the URA3 O789 O375 ccataatataaattatcagatcg taggttatggtttaatgtg O815 gcaaactagcgcaataaaggatg O662 atgtttaaatctgttgtttattcaattttagccgcttcttCGGATCCCCG GGTTAATTAA ttgtctcaatagactggcgttgtaatgagtagtgttccagGAATTCGA GCTCGTTTAAAC cgaagagcttgagcagtacaagaaacggatccccgggttaattaa Fusion primers: Amplify YFPKANMX6 to tag CgPHO4 gttttgtcttggagtatctgtagaattcgagctcgtttaaac agacgaggcttccaacgatgcaag Fusion primers: Homology to 3' end of CgPHO4 ttaattaacccggggatccgtttcttgtactgctcaagctcttcg TCT AAA AGA GTT TTC AAG GCG CCA GCG Check primer for integration of YFP GAT ATT TCA AGC downstream of CgPHO4 GGGACAACACCAGTGAATAATTCTTCACC Check primer for XFP - 3' Primers to make plasmids CGATCGGCGGCCGCGTATGCATTACCTTCTCA Amplify C. glabrata PHO4nophos with ATTAGG 5' NotI and 3' PacI restriction sites for CGACACTTAATTAATTTCTTGTACTGCTCAAG cloning into pRS313-myc13 CTCTTCG CAAATAAATATCCACTCTCGTTACAATGatccttc 1 kb CgPMU2p-PMU2 in pRS313ataaattgctattgttg myc13 accagtgaataattcttcacctttagacatTTAggatactacgatgaccg CgPMU2 stop codon 3' at gtggcggccgctctagaactagtggatcccgcgaagtcgagatgacagtg 3.6 kb (genomic clone) CgPMU2pPMU2 in pRS313-myc13 ccagtgaataattcttcacctttagacatgcaatacaccagttctgaagg CGATCGGCGGCCGCgttccttggttatcccatcg ScPHO5p-PHO5 in pRS313-myc13 O663 OD24 OD20 OD22 OD23 O98 O245 DWO5 DWO6 O213 O483 O584 O585 O480 O481 O308 locus of a C. glabrata wild-type strain Check primer for promoter-YFP integration into URA3 locus - 5' Check primer for promoter-YFP integration into URA3 locus - 3' Delete ScPHO5 with KANMX6 in DC41 O309 CGACACTTAATTAAttgtctcaatagactggcgttg ggtggcggccgctctagaactagtggatccaataacaaaatgtctaaagg vYFP-KANMX6 in polylinker of gatatcgaattcctgcagcccggggctacccgaggcaagctaaacagat pRS313-myc13 O787 c gatctgtttagcttgcctcgcaagacatatccaataggaagc O788 O318A ggtatcgataagcttgatatcgaattcctgcagcccataatataaattatca gatcg ggtggcggccgctctagaactagtggatcccacttctattactcctatcac ttg O319 ggtggcggccgctctagaactagtggatcccgaagtcgagatgacagtg c O320 ggtggcggccgctctagaactagtggatcctgataccttatgtcgctaat O430 ggtggcggccgctctagaactagtggatccATTTATCAACGAA TAACCAG Insert CgURA3 homology sequence downstream of vYFP-KANMX6 in polylinker of pRS313-myc13 Amplify 3 kB CgPMU2p - 5' (for gap repair into vYFP-KANMX6-CgURA3pRS313-myc13) Amplify 2 kB CgPMU2p - 5' (for gap repair into vYFP-KANMX6-CgURA3pRS313-myc13) Amplify 1 kB CgPMU2p - 5' (for gap repair into vYFP-KANMX6-CgURA3pRS313-myc13) Amplify 900 bp CgPMU2p - 5' (for gap repair into vYFP-KANMX6CgURA3-pRS313-myc13) O431 ggtggcggccgctctagaactagtggatccGAGAAACACTAA GCATCTAAGC O432 ggtggcggccgctctagaactagtggatccATTAAATTAATTG TAAATTAAC O433 ggtggcggccgctctagaactagtggatccATGCACTCTTTTT GAATATG O434 ggtggcggccgctctagaactagtggatccGGATAACTTGATT TTTCTTGC O435 ggtggcggccgctctagaactagtggatccGAATTTTAACAGT ACAGC O949 ggtggcggccgctctagaactagtggatccgctcagtatctttactttttaa tta O950 ggtggcggccgctctagaactagtggatccaattaaatatctcatttctcc g O951 ggtggcggccgctctagaactagtggatcccgccccgtgtatgcctcctc c O952 ggtggcggccgctctagaactagtggatcccatgtattttaaataatcaa at O1087 ggtggcggccgctctagaactagtggattatgcctcctccatgtattt O321 accagtgaataattcttcacctttagacattgtaacgagagtggatattt O320 ggtggcggccgctctagaactagtggatcctgataccttatgtcgctaat O1089 O1088 O321 O322 tttaaaatacatggaggaggcataggggcggagaaatgagat atctcatttctccgcccctatgcctcctccatgtattttaaa accagtgaataattcttcacctttagacattgtaacgagagtggatattt ggtggcggccgctctagaactagtggatcctcagcgtcatgttccccaga O429 ggtggcggccgctctagaactagtggatccGTGATTGACACA CAGAACACAG O883 ggtggcggccgctctagaactagtggatcctggtgcgctaagactactgc O323 accagtgaataattcttcacctttagacattatttcctttgttttggctt Amplify 800 bp CgPMU2p - 5' (for gap repair into vYFP-KANMX6CgURA3-pRS313-myc13) Amplify 700 bp CgPMU2p - 5' (for gap repair into vYFP-KANMX6CgURA3-pRS313-myc13) Amplify 600 bp CgPMU2p - 5' (for gap repair into vYFP-KANMX6CgURA3-pRS313-myc13) Amplify 500 bp CgPMU2p - 5' (for gap repair into vYFP-KANMX6CgURA3-pRS313-myc13) Amplify 400 bp CgPMU2p - 5' (for gap repair into vYFP-KANMX6CgURA3-pRS313-myc13) Amplify 780 bp CgPMU2p - 5' (for gap repair into vYFP-KANMX6CgURA3-pRS313-myc13) Amplify 760 bp CgPMU2p - 5' (for gap repair into vYFP-KANMX6CgURA3-pRS313-myc13) Amplify 740 bp CgPMU2p - 5' (for gap repair into vYFP-KANMX6CgURA3-pRS313-myc13) Amplify 720 bp CgPMU2p - 5' (for gap repair into vYFP-KANMX6CgURA3-pRS313-myc13) Amplify 730 bp CgPMU2p - 5' (for gap repair into vYFP-KANMX6CgURA3-pRS313-myc13) Amplify at start codon CgPMU2p - 3' (for gap repair into vYFP-KANMX6CgURA3-pRS313-myc13) Fusion PCR primers to mutate CCCGTG (to delete GTG) in 1 kb CgPMU2p-YFP Amplify 1 kB CgPHO84p - 5' (for gap repair into vYFP-KANMX6CgURA3-pRS313-myc13) Amplify 400 bp CgPHO84p - 5' (for gap repair into vYFP-KANMX6CgURA3-pRS313-myc13) Amplify 300 bp CgPHO84p - 5' (for gap repair into vYFP-KANMX6CgURA3-pRS313-myc13) Amplify at start codon CgPHO84p 3' (for gap repair into vYFPKANMX6-CgURA3-pRS313-myc13) O429 O995 O994 O323 O75 O76 O78 O79a O664 O641 O155 O156 O153 O154 O151 O152 O1207 O1208 O1191 O1192 O1193 O1194 O1201a O1202a O1203 O1204 O1205 O1206 O1211 O1212 O1225 O1226 O1221 O1222 O1217 O1218 O1229 O1230 ggtggcggccgctctagaactagtggatccGTGATTGACACA CAGAACACAG gtattagattaggaccaGaGCtCcatgtgggcccctctttc gaaagaggggcccacatgGaGCtCtggtcctaatctaatac accagtgaataattcttcacctttagacattatttcctttgttttggctt qPCR primers gaccaaactacttacaactcc ccactttcgtcgtattcttgcttg gccttgggtactctgatcgacc gtgctgtcggcgacagtaac gtgatgttactgttcattttgattatcctg gatacatcgatcacagtgtaaactag TCCTACGAACTTCCAGATGGT GGCAGATTCCAAACCCAAAA GCCGGTTCCAAAAACGTTTA GACAGTGAAGACGGATACCCA TGCTTGTAACTCATGTCCTGC TTGAGGTCAAGTTCAAACCCT ChIP primers Fusion PCR primers to mutate CACGTG (ScPho4 binding site) in 400 bp CgPHO84p-YFP acggcccatcagcagcagcttc cttcatagcacagagctcatagc tgaagacactgagagcacgcg atcacatttccactgcggggc aatccatataagtgggaagcg aggtccccgcagaattctccg ctgctatactgctaagactaag tagattaggaccacacgtgc tgtgcggaaagaggggcccac caaattttttttccctgcatc agctcatctctaatatttccaat agcaacttggatccggtagtt gcgtgcggtaaagtcggtgcc aaccaacagttcttaggcttgc gcatagctatttcaatgtcttc ttcttaaagaactagggaggcac acatcatactaagatcaaaaaag aggaggcatacacggggcggag tctcacgtatagctgagattg acattattgtttatgcatgtc -950 bp CgPHO84 promoter AGATAAGTTTTTCGTTTCACCAT ATGAACAGTGGAATTTGCTGGC CgACT1 5' qPCR CgACT1 3' qPCR CgPHO84 5' qPCR CgPHO84 3' qPCR CgPMU2 5' qPCR CgPMU2 3' qPCR ScACT1 5' qPCR ScACT1 5' qPCR ScPHO84 5' qPCR ScPHO84 3' qPCR ScPHO5 5' qPCR ScPHO5 3' qPCR -450 bp CgPHO84 promoter -400 bp CgPHO84 promoter -200 bp CgPHO84 promoter -150 bp CgPHO84 promoter -100 bp CgPHO84 promoter +1500 bp CgPHO84 promoter -2333 bp CgPMU2 promoter (CACGTG) -800 bp CgPMU2 promoter -500 bp CgPMU2 promoter +500 bp CgPMU2