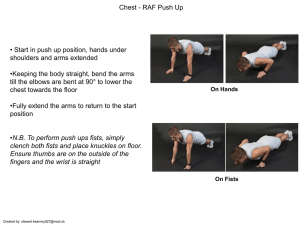
an assessment of shallow and mesophotic reef brachyuran crab
advertisement

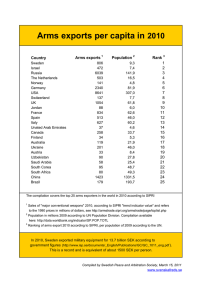
Electronic Supplementary Material AN ASSESSMENT OF SHALLOW AND MESOPHOTIC REEF BRACHYURAN CRAB ASSEMBLAGES ON THE SOUTH SHORE OF OʻAHU, HAWAIʻI KKC Hurley, MA Timmers, LS Godwin, JM Copus, DJ Skillings, RJ Toonen Table S1 Number of brachyuran crabs at different depths, ARMS units pooled among sites at each depth. Depth Number of ARMS units 12 m 9 30 m 6 60 m 6 90 m 6 Total number of crabs 380 100 98 85 Mean SD SE Observed morphospecies Unique morphospecies 42 25 24 21 27 4 5 6 42.22 11.35 3.78 16.67 8.45 3.45 16.33 6.15 2.51 14.17 6.64 2.71 Table S2 Number of brachyuran crabs at different depths, ARMS units pooled among sites at each depth, but with the most common brachyuran (Chlorodiella spp.) excluded from the analyses. Depth Number of ARMS units 12 m 30 m 60 m 90 m 9 6 6 6 Total number of crabs 154 74 75 77 Mean SD SE 17.11 12.33 12.50 12.83 6.66 6.02 5.28 7.35 2.22 2.45 2.15 3.00 Table S3. List of taxa identified in autonomous reef monitoring structures (ARMS) deployed at 12, 30, 60, and 90 m. All bold text is family names, unless otherwise indicated. Total counts by family are within parenthesis. *Morphospecies names are included. Acidopsidae (3) Acidopsidae sp2010* Parapilumnus sp2049* Aphanodactylidae (1) Aphanodactylus sp502* Catoptridae (17) Catoptrus inequalis (Rathbun, 1906) Domeciidae (7) Domeciidae sp1* Domecia glabra Alcock, 1899 Dynomenidae (29) Dynomene hispida (Latreille, in Milbert, 1812) Epialtidae (40) Hyastenus sp1* Perinia tumida Dana, 1851 Leucosiidae (6) Heteronucia sp1* Nucia sp1* Majidae (1) Schizophroida sp2011* Palicidae (8) Exopalicus maculatus (Edmondson, 1930) Parthenopidae (3) Daldorfia horrida (Linnaeus, 1758) Percnidae (8) Percnon abbreviatum (Dana, 1851) Pilumnidae (58) Pilumnus acutifrons Rathbun, 1906 Pilumnus nuttingi Rathbun, 1906 Pilumnus oahuensis Edmondson, 1931 Pilumnus sp259* Pilumnus sp364* Pilumnidae sp2018* Pilumnus taeniola Rathbun 1906 Portunidae (46) Carupa tenuipes Dana, 1852 Laleonectes nipponensis (Sakai, 1938) Thalamita coeruleipes Hombron & Jacquinot, 1846 Thalamita admete (Herbst, 1803) Thalamita bevisi (Stebbing, 1921) Thalamita seurati Nobili, 1906 Thalamita sp124* Thalamita sp308* Thalamita sp2042* Thalamita sp3026* Thalamita sp3011* Thalamitoides quadridens A. Milne-Edwards, 1869 Trapeziidae (4) Trapezia flavopunctata Eydoux & Souleyet, 1842 Trapezia intermedia Miers, 1886 Xanthidae (417) Chlorodiella sp12* Epiactaea nodulosa (White, 1848) Garthiella aberrans (Rathbun, 1906) Juxtaxanthias intonsus (Randall, 1840) Liocarpilodes integerrimus (Dana, 1852) Liocarpilodes sp1071* Liomera bella (Dana, 1852) Liomera medipacifica (Edmondson, 1951) Liomera tristis (Dana, 1852) Lophozozymus dodone (Herbst, 1801) Lophozozymus sp439* Lybia edmondsoni Takeda & Miyake, 1970 Medaeus ornatus Dana, 1852 Olenothus sp551* Paractaea rufopunctata (H. Milne Edwards, 1834) Paramedaeus simplex (A. Milne-Edwards, 1873) Pilodius areolatus (H. Milne Edwards, 1834) Pilodius flavus Rathbun, 1894 Platypodia semigranosa (Heller, 1861) Platypodia sp1* Platypodia sp3014* Pseudoliomera sp1068* Pseudoliomera variolosa (Borradaile, 1902) Tweedieia laysani (Rathbun, 1906) Xanthias canaliculatus Rathbun, 1906 Xanthias latifrons (de Man, 1887) Xanthidae sp1* Xanthidae sp1027* Superfamily Majoidea (14) Majoidea sp261* Majoidea sp1032* Majoidea sp2051* Majoidea sp3027* Majoidea sp3084* Table S4 Permutational analysis of variance (PERMANOVA) and permutational analysis of dispersion (PERMDISP) p-values, obtained using 9999 permutations, for pairwise brachyuran crab assemblage comparisons between depths of 12, 30, 60, and 90 m. Resemblance matrices are based on the Jaccard index. PERMANOVA PERMDISP Depth (m) comparisons P-value Unique permutations P-value 12, 30 0.0003 4300 0.4484 12, 60 0.0003 4342 0.6933 12, 90 0.0002 4291 0.5687 30, 60 0.0017 462 0.7163 30, 90 0.0022 462 0.915 60, 90 0.0023 461 0.8227 Table S5 Permutational analysis of variance (PERMANOVA) and permutational analysis of dispersion (PERMDISP) p-values for pairwise comparison of brachyuran crab assemblages between 12, 30, 60, and 90 m. To balance uneven sampling design, three of nine ARMS units were randomly removed from 12 m set before tests (all other depths had 6 ARMS units). PERMANOVA PERMDISP Depth (m) comparisons P-value Unique permutations P-value 12, 30 0.002 460 0.4328 12, 60 0.024 461 0.1856 12, 90 0.0021 462 0.6033 30, 60 0.0016 462 0.7171 30, 90 0.002 462 0.8913 60, 90 0.0028 462 0.6355 Fig. S1 Abundance of crabs in individual ARMS units at each depth in this study. Fig. S2 Mean number of brachyuran crabs (± SE) in the five most common families recovered from each ARMS unit by depth. Note the significant decrease in abundance of crabs (particularly Xanthidae) from shallow to mesophotic depths. Fig. S3 Box and whisker plot of the number of brachyuran crabs recovered by family pooled across all ARMS units in this study. Dark lines represent the median. The boundaries of the boxes are the first and third quartile of the data. Whiskers represent the maximum and minimum value at each depth and circles represent outliers. Fig. S4 Box and whisker plot of the number of brachyuran crabs recovered from each ARMS unit by depth (12, 30, 60 or 90 m). The dark line is the median number of individuals per ARMS unit, the box contains the middle two quartiles, whiskers represent the maximum and minimum numbers of individuals recorded from any ARMS unit at that depth and circles represent outliers. Fig. S5 Box and whisker plot of the number of brachyuran crabs recovered from each ARMS unit at four depths (12, 30, 60 or 90 m) excluding Chlorodiella spp. The dark line is the median number of individuals per ARMS unit, whiskers represent the maximum and minimum numbers of individuals recorded from any ARMS unit at that depth and circles represent outliers. Fig. S6 Mean number of brachyuran crabs (± SE) in the five most common families recovered from each ARMS unit by depth, but with Chlorodiella spp. excluded. There is a general decrease in abundance of crabs from shallow to mesophotic depths in other families is retained, but is reversed in the Xanthidae. Fig. S7 Box and whisker plot of the number of brachyuran crabs (excluding Chlorodiella spp.) recovered by family pooled across all ARS units in this study. Dark lines represent the median. The boundaries of the boxes are the first and third quartile of the data. Whiskers represent the maximum and minimum value at each depth and circles represent outliers. Fig. S8 Canonical analysis of principal coordinates (CAP) ordination of brachyuran crab assemblages (excluding Chlorodiella spp.) in autonomous reef monitoring structures (ARMS) at at 12 m (green crosses), 30 m (orange triangles), 60 m (blue squares) and 90 m (red diamonds). This figure supplements Fig. 4 that includes all brachyuran species. Resemblance matrix based on Bray– Curtis similarity; Spearman rank restricted to ρ = 0.8 Figure S9 Vector overlays of Spearman rank correlations of individual crab species on canonical analysis of principal coordinates (CAP) ordination of brachyuran crab assemblages for 24 autonomous reef monitoring structures (ARMS) at 12, 30, 60, and 90 m (resemblance matrix based on Bray-Curtis similarity; Spearman rank restricted to ρ = 0.8). Three ARMS were randomly selected to be removed from 12 m to account for the uneven sampling design (6 units at 30, 60, and 90 m). Figure S10 Rarefaction curves with 95% confidence intervals for crab assemblages from (a) 12 m, (b) 30 m, (c) 60 m, and (d) 90 m. For 12 m, the number of ARMS used as reference is 9 units. For all other depths, reference sample = 6.