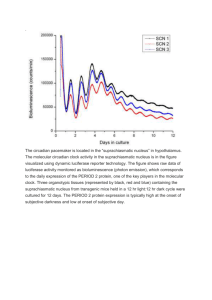
P155
advertisement

P155 Modeling the circadian oscillator protein network in Drosophila melanogaster Vu Lam , Ying Li , Jonathan Diehl , Joanna Chiu 1 1 1 1 1University of California, Davis, Davis, CA, UNITED STATES Organisms, from bacteria to mammals, rely on the circadian clock for timekeeping and orchestrating daily rhythms of physiology and behavior to coordinate with the environment and maximize survival. Central oscillator function relies on dynamic and temporal changes in the circadian interactome centered around a number of core transcriptional regulators. In Drosophila melanogaster, these core proteins are PERIOD (PER), TIMELESS (TIM), CLOCK (CLK), and CYCLE (CYC). Temporal changes in proteinprotein interactions (PPIs) between PER, TIM, CLK, and CYC and other cellular complexes, such as those responsible for light entrainment, protein posttranslational modifications, proteasome degradation, localization, and chromatin remodeling, directly regulate the cycling of clock protein abundance and function, and determine the timing for which they are active as regulators of the circadian transcriptome. To better understand the interface between the oscillator and cellular protein complexes, we modeled the complex design of the Drosophila circadian PPI networks by establishing cellular interactomes of PER, TIM, CLK, and CYC using affinity purification (AP) and label-free quantitative mass spectrometry (MS) and generated protein networks centering around these core circadian proteins in Drosophila S2 cell nucleus and cytoplasm. Furthermore, we developed a data analysis pipeline to model AP-MS protein network data and annotate the resulting protein networks by incorporating published PPIs, knowledge about CRAPome, i.e. common AP-MS contaminants, as well as coexpression data in circadian clock neurons. Our circadian oscillator PPI networks, together with characterization of PPI candidates will be presented.