CyberSEES_Data_Management_Plan_2015Final
advertisement

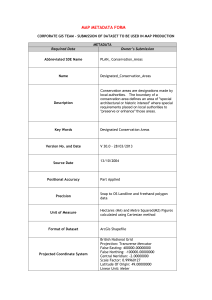
Data Management Plan 1 Overview Title: CyberSEES Marine Biodiversity Virtual Laboratory project data management plan; Author: S. Beaulieu, WHOI; Date: 20150219; Revision: not applicable. Proposal submitted to NSF Solicitation 15524 2 Expected Data 2.1 Data: This project does not generate new observational data. This project uses observational data that are already in the public domain, from three main sources: 1) prokaryotic and eukaryotic DNA sequence data from Visualization and Analysis of Microbial Population Structures (VAMPS; http://vamps.mbl.edu/), 2) phytoplankton imagery data from Imaging FlowCytobot (IFCB; http://ifcbdata.whoi.edu/), and 3) environmental data from the Martha’s Vineyard Coastal Observatory (http://www.whoi.edu/mvco/data). Derived datasets and data products, model data, and metadata for these, will be generated and managed by this project. 2.2 Data Formats: The derived data generated in this project will be in non-proprietary formats (e.g., csv, NetCDF, JSON, RDF). Open source software can be used to read the data. 2.3 Data Generation & Acquisition: The derived and model data will be generated with open source software (i.e., IPython Notebook) and initially stored on local servers. We will consider the quality control and standards already applied to the data we are pulling into our product workflows from other sources. Our derived data will be generated on demand from source data that are available at different frequencies, ranging from near real-time (IFCB data) to monthly (DNA sequence data). We will generate derived datasets and metadata at intermittent frequencies during the prototyping phase of this project. Ultimately, since the biodiversity indicators will be produced from aggregated source data, the frequency will be constrained by the resolution of the included source datasets. 2.4 Software: Three types of software development are involved in this project: 1) software for the IFCB Dashboard to be shared with our industry partner, McLane Research Labs, 2) scripting for data integration, including production of biodiversity indicators, analysis and modeling (e.g., IPython Notebooks), and 3) ontology development for reusable semantic structures. All software in this project will be managed in a code repository supporting change-tracking and revision control (e.g., GitHub, svn) accessible to co-PIs and project partners during the project period, and will be transitioned to public availability once documented versions are ready. For types 1 & 2, we will consider an official OSF license such as LGPL or the Apache license (http://opensource.org/licenses). For 3, we will consider a Creative Commons Attribution-ShareAlike 3.0 Unported License (http://creativecommons.org/licenses/by-sa/3.0/). 2.5 Documentation and Metadata: Data and metadata will conform to national and international standards, e.g., taxonomic standards (microbes: Global Alignment for Sequence Taxonomy with ARBSILVA; eukaryotes: WoRMS, which is used by Ocean Biogeographic Information System), W3C standards (i.e., RDF, OWL). For metadata, we will use schema terminology and biological data definitions as specified by U.S. IOOS Biological Observations project (i.e., Darwin Core and IOOS vocabularies, CF conventions). Metadata will be generated automatically when possible, manually otherwise. Metadata in RDF will be stored as a triple store database such as Virtuoso Universal Server (http://virtuoso.openlinksw.com/). Metadata provided to repositories/archives with derived datasets will be in the respective required formats. We expect to use W3C ontologies (i.e., PROV, DCAT) and to adopt from ontologies developed at RPI TWC (i.e., ECO-OP, GCIS). RPI provides a dataset identifier service (via Renssalaer Data Services). For dataset identification, we will reuse existing URIs and URLs as appropriate (e.g., DOIs), following the Linked Open Data approach; where it is necessary to mint unique identifiers we will attempt to reuse local identifiers (e.g., filenames) and apply namespace scoping following XML and RDF best practices. 3 Data Storage and Preservation 3.1 Storage and Backup During the Project: Co-PIs are responsible for data storage and backups for their respective components of the project. All final products, some intermediate products, metadata, and code will be stored. There are no non-digital data in this project. Derived datasets and data products will This work is licensed under the Creative Commons Attribution-ShareAlike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-sa/3.0/ or send a letter to Creative Commons, 171 Second Street, Suite 300, San Francisco, California, 94105, USA. be stored on local servers at our respective institutions, while code may be stored on third-party servers with online access. Access controls may include login during the project period, with public availability at the conclusion of the project. Local storage and backups per institution are described in detail in Facilities statements. 3.2 Data Capacity & Volume: Although the volume of data ingested into the product workflows may be large (e.g., genetic sequence data and IFCB imagery data), the derived and model datasets are expected to be relatively low-volume, tabular data (e.g., csv files from <1 to 100MB). For frequency, see 2.3. 3.3 Security: Not applicable at this time. 3.4 Operation Storage Post Project Completion: RPI via Rensselaer Data Services (http://data.rpi.edu/), takes the preservation role and responsibility for data post-completion. 3.5 Long Term Archiving and Preservation: Relevant derived data and required metadata will be provided to the respective source data repositories for archiving (i.e., VAMPS). We will most likely choose NSF’s Biological and Chemical Oceanography Data Management Office (BCO-DMO; http://www.bcodmo.org/) as repository for derived data from the IFCB, as BCO-DMO provides copy to NODC for archiving. 3.6 Roles and Responsibilities: Lead PI Fox makes decisions regarding the overall data management. CoPIs and Senior Personnel make decisions regarding day-to-day data management. Guidance from respective repositories/archives (3.5) for required metadata will be obtained by PI and Co-PIs. 4 Data Retention Source data and intermediate data products will be shared among co-PIs and project partners during the project period. Final data products will be shared with open access post-project. The data lifecycle will encompass: access store QA/QC produce products preserve (with a loop between "produce products" and "store" in which we will incorporate versioning. No embargo periods. See also 3.5. 4.1 Operational Data: Lead PI Fox at RPI (via data.rpi.edu) takes responsibility for the data in the nearterm following project completion. 4.2 Archival Data: Co-PI Beaulieu will provide derived data to respective data archives. 5 Data Sharing and Dissemination Derived datasets and data products, model data, and their metadata, for biodiversity in the Northeast Shelf Large Marine Ecosystem will be shared publicly once quality control and metadata have been applied and no later than the end of the project period. Data will be available through local servers in the nearterm and through community repositories (e.g., VAMPS, BCO-DMO) and archives (e.g., NODC) in the long-term. 5.1 Stakeholders: Data will be shared regionally (e.g., NOAA Northeast Fisheries Science Center) and will be disseminated to other national (e.g, NASA) and international (e.g., ICES WGNARS working group, IMBER, GOOS, CBD) stakeholders. 5.2 Privacy and Confidentiality: Not applicable. 5.3 Ownership, Copyright and IP: There will be no copyright on data. All IP (data and software) ownership will be based on originator policies. Materials generated by an individual will be owned by the individual (and/or their organization). All joint works will be jointly owned. 5.4 Third Party Data: All of the source data will be obtained from publicly available datasets, most of which were contributed by co-PIs. Any third party data, e.g. from the Web, will be used in accordance with accompanying licensing and given suitable attribution. We will respect all conditions on use, sharing, and re-dissemination. Also, see 5.7. 5.5 Legal and Regulatory: No regulatory constraints on sharing and dissemination of data. 5.6 Re-Use: Re-use of the data is strongly encouraged. Citations are requested in conformance to the ESIP guidelines, http://wiki.esipfed.org/index.php/Interagency_Data_Stewardship/Citations. 5.7 Ethical Requirements: We will adhere to any intellectual property requirements of the providers of source data that we bring into the product workflows. This work is licensed under the Creative Commons Attribution-ShareAlike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-sa/3.0/ or send a letter to Creative Commons, 171 Second Street, Suite 300, San Francisco, California, 94105, USA.