pmic12191-sup-0002-text
advertisement

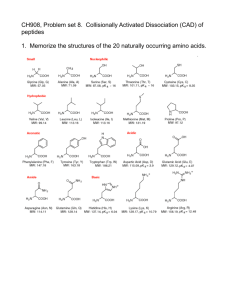
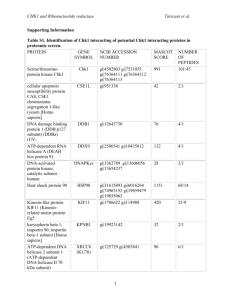
Supporting Information 2 – Materials and Methods Corneal tissue – A corneal flap consisting of epithelium, Bowman’s layer, and superficial stroma was removed from GCD2 post-LASIK patients having LASIK surgery again as treatment for protein aggregation. Control tissue consisting of superficial stroma was excised from the cornea during ReLEx SMILE surgery of healthy individuals. Isolated epithelium was obtained post-mortem from a 96-year-old male with no history of corneal defects. All tissues were kept at -80°C until further use. The patient and control groups consisted of six biological samples each, representing mixed genders and with an average age of 34 years and 36 years, respectively (Supporting Information 1). The tissue was obtained and used for scientific purpose and ethical issues have been handled according to Danish and South Korean healthcare laws. All work was carried out in accordance with the Declaration of Helsinki. Sample preparation – GCD2 patient and control tissues were lyophilized for 2 h using a speedvac concentrator and grinded into powder using liquid nitrogen. Approximately, 0.3mg of each tissue was subjected to 0.66 M CNBr in 70% trifluoroacetic acid at 23°C. The following day the tissues were lyophilized and re-suspended in 8 M Urea in 0.2 M Tris-HCl, pH 8.3, reduced in 5 mM dithiothreitol for 30 min, and alkylated in 15 mM iodoacetamide for 30 min. Samples were then diluted 5 times using 0.1M Tris-HCl, pH 8.3, followed by overnight digestion with 1:50 w/w sequencing grade modified trypsin (Sigma) at 37°C. The samples were desalted using POROS 50 R2 RP column material (Applied Biosystems) packed in GELoader Tips (Eppendorf). LC-MS/MS – LC-MS/MS analyses were performed on an EASY-nLC II system (Thermo Fisher Scientific) connected to a TripleTOF 5600+ mass spectrometer (AB SCIEX) equipped with a NanoSpray III source (AB SCIEX) and operated under Analyst TF 1.6.0 control. The CNBr and trypsin cleaved samples were dissolved in 0.1% formic acid, injected and trapped on an in-house packed trap column (2 cm x 100 μm I.D) using RP ReproSil-Pur C18-AQ 3 μm resin (Dr. Maisch GmbH). Peptides were eluted from the trap column and separated on a 15 cm analytical column (75 μm i.d.) pulled and packed inhouse with RP ReproSil-Pur C18-AQ 3 μm resin (Dr. Maisch GmbH) and sprayed directly into the mass spectrometer. Peptides were eluted at a flow rate of 250 nL/min using a 20 min gradient from 5% to 35% phase B (0.1% formic acid and 90% acetonitrile). The acquisition method used for the area based extracted ion chromatogram (XIC) quantification was set up as an information-dependent acquisition (IDA) experiment collecting up to 25 MS/MS spectra in each 1.6 sec cycle using an exclusion window of 6 sec. The GCD2 patient and control samples were run in triplicates on the mass spectrometer. Data processing – For protein identification all raw MS files were processed using Mascot Distiller 2.5.1.0 (Matrix Science) and MGF files generated were search in the Swiss-Prot (v. 2015_03) Homo sapiens database using Mascot 2.5.0 (Matrix science, London, UK). CNBr+Trypsin was selected as the digestion enzyme allowing one missed cleavage. To address endogenous P1 cleavage sites a second search was performed using semi-trypsin as specified enzyme. In both searches carbamidomethyl was entered as a fixed modification, whereas hydroxylation of proline, oxidation of methionine, and C-terminal conversion of methionine to homoserine and homoserine lactone by CNBr were used as variable modifications. All data were searched with a mass tolerance of the precursor and product ions of 10 ppm and 0.2 Da using ESI-QUAD-TOF as the instrument setting and all searches was adjusted to a 1% FDR at the peptide level. All data were imported and processed using MS Data Miner v. 1.3.0 [1]. All protein identifications based on at least one peptide with an ion score cut-off of 45 or minimum two peptides with an ion score cut-off of 30 was included for further data analysis (Supporting Information 3). Protein identifications based on one peptide was subjected to manual validation before accepted (Supporting Information 5). To address endogenous P1 cleavage sites all TGFBIp peptide spectra with an ion score cut-off of minimum 45 within the semi-tryptic search was extracted and the distribution of P1 amino acid residues of the non-tryptic cleavage site was calculated and compared between control and patient groups (Supporting Information 9 and 11). For the area-based XIC quantification Mascot Distiller 2.5.10 was used and the mascot search was performed using the same settings as for protein identification above except that the default average [MD] quantitation protocol was selected using a 1% FDR threshold at the peptide level, number of peptides used for quantitation was 3, matched rho was 0.8, XIC threshold was 0.3 and isolated precursor threshold was set at 0.7. The area-based XIC quantification was based on the average intensity of the 3 most abundant peptides per protein and displayed as a molar percentage by dividing the average intensity with the total intensity of the given MS analysis (Supporting Information 4). Using Gene Ontology annotations all extracellular proteins were ranked according to their molar fraction of a sample and the average molar percentage across patient and control groups were compared if a protein was quantified in minimum 3 of the 6 biological replicates in a group (Table 1, Supporting Information 8). 2D-PAGE – Corneal powder was dissolved in lysis buffer (7 M urea, 2 M thiourea, 4 % (w/v) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 10 mM Tris pH 8.8, 5 mM EDTA, cOmplete EDTA-free protease inhibitor cocktail (Roche Diagnostics, Basel, Switzerland), 0.5 % (v/v) IPG buffer of pH 3-10 or 4-7 and 10 mM DTT) and incubated rotating for 1h at room temperature. The samples were centrifuged and supernatants were assayed for protein content using the 2D Quant kit (GE Healthcare Life Sciences, Buckinghamshire, England). The protein amount was verified and adjusted on a 1D SDS-PAGE gel. For each sample 2 µg of total protein was loaded onto 7-cm Immobiline DryStrips (GE Healthcare Life Sciences) with pH range 3-10 or 47. The strips were rehydrated overnight at room temperature and isoelectric focusing was performed using the Ettan IPGphor System II (GE Healthcare Life Sciences) for 5.3 kVh. Following isoelectric focusing proteins were reduced in equilibration buffer (50 mM Tris-HCl pH 8.8, 6 M urea, 30 % (v/v) glycerol and 2 % SDS) with 6.5 mM DTT and then alkylated in equilibration buffer containing 10 mM iodoacetamide. The strips were placed on top of 10 % SDS-polyacrylamide gels and the second-dimension were run at 17.5 mA per gel in running buffer (25 mM Tris, 192 mM glycine and 0.1 % (w/v) SDS). Immunoblotting of TGFBIp – Proteins separated by 2DE were electroblotted onto a polyvinylidene difluoride (PVDF) membrane (Immobilon-P, Millipore, Billerica, MA) for immunoblotting. The membranes were blocked in 40 ml 5 % dry milk solution in TBS-T (20 mM Tris-HCl pH 7.6, 137 mM NaCl and 0.1 % tween) for 1 h at room temperature. An antiserum from rabbit against rhTGFBIp [2] was added to the blocking solution in a ratio of 1:10000 and membranes were incubated overnight at 4°C. The membranes were washed thoroughly with TBS-T, before incubated for 2 h at room temperature in TBS-T containing goat anti-rabbit IgG peroxidase conjugate (Sigma-Aldrich Co., St. Louis, MO) in a ratio of 1:10000 in 5 % milk. Finally, the membranes were washed thoroughly with TBS-T and developed for 0.25-1 min using the enhanced chemiluminescence (ECL) Western blotting detection system and reagents (GE Healthcare Life Sciences). References [1] Dyrlund, T.F., Poulsen, E.T., Scavenius, C., Sanggaard, K.W., Enghild, J.J., MS Data Miner: a web-based software tool to analyze, compare, and share mass spectrometry protein identifications. Proteomics 2012, 12, 2792–2796. [2] Runager, K., Klintworth, G.K., Karring, H., Enghild, J.J., The insoluble TGFBIp fraction of the cornea is covalently linked via a disulfide bond to type XII collagen. Biochemistry 2013, 52, 2821–2827.