Glossary, ESM - Springer Static Content Server
advertisement

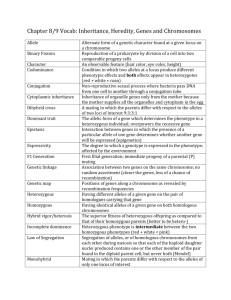
Glossary, ESM function as start and stop codons, flanking the coding stretch on the messenger RNA. allele One of two or more alternate versions of a gene at the same location (locus) along a chromosome; different alleles can be due to mutations. Alleles contribute to the exhibited phenotype. Humans are diploid organisms having two alleles at each genetic locus, with one allele received from each parent. Genotypes are homozygous if there are two identical alleles at a particular locus and heterozygous if the two alleles differ. degeneracy Gene networks exploit alternate strategies to preserve the output ; they exemplify degeneracy, by adjusting expression of different genes and using multiple pathway combinations to achieve functional plasticity, producing a similar phenotypic outcome in one context and dissimilar ones in other contexts. Degeneracy is a key organizational feature of our genetic code; it greatly increases the capacity of a limited and fixed number of genes to yield remarkable morphomechanical complexities. allelic heterogeneity Different mutations in the same gene at the same chromosomal locus that cause a single phenotype. alternative splicing The processing of an RNA transcript into different mRNA molecules. Our genes are usually organized as a series of alternating exons that encode amino acids and introns that do not. After the RNA transcript is synthesized, RNA splicing enzymes remove the intron sequences and attach the exon sequences together to produce the final mRNA molecule. Sometimes an exon sequence is treated as an intron and is removed, producing an alternative form of the mRNA. For many DNA sequences alternative splicing patterns may occur under different circumstances (in different cells, tissues, and organs), thus giving rise to diverse proteins from the same DNA stretch. assembly Computational reconstruction of a longer DNA/RNA sequence from smaller sequence reads bioinformatics The study of collecting, sorting, and analyzing DNA and protein sequence information using computers and statistical techniques. chromatin Complex of DNA, RNA, histones, and non-histone proteins from which eukaryotic chromosomes are formed. codon Each of the different nucleotide triplets in DNA that, when transcribed into RNA, are then translated into an amino acid in a protein. Since any one of the four kinds of nucleotides of which a DNA strand is built can occupy each of the three positions of a codon, there is a totality of 43 = 64 different codons as compared to only 20 available amino acids; therefore, most amino acids are encoded by more than one codon (the genetic code is redundant) and most DNA sequence changes affecting the third position in a codon do not change the resulting protein. Some (noncoding) codons DNA sequencing Processes determining the exact order of the base pairs in a segment of DNA. epigenetic inheritance (epigenetics) Inheritance of a trait for one or more cell generations that is not transmitted by the genetic material proper and is reversible. A case of epigenetic inheritance is the transmission of DNA methylation patterns. epistasis Gene interaction and, particularly, interaction between different alleles at different genes. Epistasis is a phenomenon that consists of the effect of one gene being dependent on the presence of one or more modifier genes (genetic background), and can occur at the same step or at different stages of a pathway or process. epistatic interaction The influence of the interaction of multiple loci on phenotypic variation.. euchromatin State of chromatin in which the DNA is partially or fully uncoiled and transcriptionally active, in contrast to the inert heterochromatin. exons Rather than forming a continuous stretch of coding DNA, a gene may consist of a chain of coding and noncoding sequences called exons and introns, respectively. The RNA transcript of such a gene goes through a process of splicing in which the introns are excised yielding a "mature" messenger RNA composed only of exons; each exon codes for a specific portion of the complete protein. gene expression General process by which the information encoded in a gene is converted into an observable phenotype, such as the production of proteins. It pertains to DNA transcription into RNA, RNA splicing/edition processes, translation, and post-translational protein modifications. gene network A graphical illustration for exploring the functional interactive linkages and the potential coordinate regulations of genes. Networks consist of ‘‘nodes,’’ the genes and their regulators, joined together by ‘‘edges,’’ which represent physical and/or regulatory interactions. genome All the genetic material in the chromosome set of a particular organism. In diploid organisms there are two genomic complements, one complement inherited from each parent. genotype The combination of alleles on the two chromosomes of an individual, representing the inherited genetic makeup or genetic constitution of a cell or organism, which is modulated by the environment before being expressed as a phenotype. haplotype A set of allelic states, or a series of polymorphisms, found at neighboring loci in a chromosome and tending to be inherited jointly. A haplotype shared among unrelated individuals affected with a genetic disease may indicate that a gene causing the disease maps to that genomic region. heterochromatin Zones of chromatin with DNA that remains condensed and transcriptionally inactive or repressed. homologous (or homolog) A gene or protein inherited in two or more species by a common ancestor. While homologous genes can be similar in sequence, similar sequences are not inevitably homologous. Human Genome Project (HGP) An international research project begun in 1990 and completed in 2003 that mapped and sequenced the three billion DNA bases of the human genome. monogenic (single gene) alterations. However, for some phenotypes it is becoming clear that mutations in a single gene can be insufficient to cause a specific phenotype without sequence alterations in other genes. Further variation in phenotype or penetrance can be caused by loss or alteration of contiguous genes. Modifier genes have become increasingly recognized as an important source of phenotypic variation that may explicate the complex relationship of phenotype to genotype. monogenic Involving a single gene. Monogenic diseases are attributable to mutations in a single gene; the defective gene is inherited from a parent in most instances, but may also arise from spontaneous mutation. multifactorial Traits/syndromes that are determined by a combination of genetic as well as non-genetic factors, each contributing to the overall phenotype. mutation A gene mutation is a permanent alteration in the DNA sequence that makes up a gene, such that the sequence differs from what is found in most people. Mutations range in size; they can affect anywhere from a single DNA building block (base pair) to a large segment of a chromosome that includes multiple genes. Next Generation Sequencing (NGS) Highthroughput DNA and RNA (coding and non-coding) sequencing methods which parallelize massively the sequencing process, producing thousands or millions of sequences at once; they can explore gene expression changes, epigenomic pattern, etc, on a genome-wide scale and with single base precision. introns Rather than forming a continuous stretch of coding DNA, a gene may consist of a chain of noncoding and coding sequences called introns and exons, respectively. The RNA transcript of such a gene goes through a process of splicing in which the introns are excised yielding a "mature" messenger RNA composed only of exons. The introns may serve other functions, such as regulation of the splicing and translation processes, or even may be exons of other genes. penetrance The likelihood, or probability, that a particular genotype will be expressed in the phenotype. A penetrance of 100% means that the associated phenotype always occurs when the corresponding genotype is present; similarly, if only 25% of those carrying a particular allele (such as a disease-causing mutation) exhibit a phenotype (the disease), the penetrance is 25%. locus (pl. loci) In genetics, a specific site of a gene on a chromosome. All the alleles of a particular gene occupy the same locus. phenotype Expressed morphomechanical traits or other characteristics, including the presence or absence of a disease, regardless of whether or to what extent the traits are the result of genotype or environment, or of the interaction of both. modifier genes For many diseases, including various genetic cardiomyopathies, no clear relationship of phenotype to genotype can be demonstrated. Most publications pertaining to the genetics of pleiotropic syndromes have resulted from the study of cases involving high-penetrance pleiotropy (from the Gk. pleio and tropic, indicating "more" and “forms, respectively.") A situation in which mutations in one single gene can bring forth various morphomechanical phenotypic traits/forms. polygenic trait (multifactorial trait) Any phenotype that results from the effect of multiple genes at two or more loci, with possible modulating environmental influences too. Cardiovascular examples include hypertension, heart failure, hypercholesterolemia, etc. polymorphism Genome segment (locus), within or outside a gene, in which alternate forms (alleles) are present, having arisen from mutational events. A genetic variation is polymorphic if all alleles are found at frequencies >1% in the general population. In clinical genetics, a polymorphism refers to any genetic variation that, in contrast with a mutation, is not known to be a direct cause of disease. However, the distinction may be fuzzy, as the path from genetic variation to disease can sometimes be complicated. remodeling The (mal)adaptive change of tissue and organ morphology due to external or internal stimuli. single-nucleotide polymorphism (SNP) Variation in a genetic sequence that affects only one of the basic nucleotide building blocks—adenine, guanine, thymine, or cytosine—in a segment of a DNA molecule and occurs in more than 1 percent of a population. transcription The process by which RNA polymerase assembles an RNA molecule complementary to the template strand of the DNA. The product may be messenger RNA, transfer RNA or ribosomal RNA. translation The process by which messenger RNA directs a ribosome to assemble a polypeptide. As specified in the genetic code each codon in the mRNA corresponds to an amino acid. The assembly of the protein stops when the ribosome encounters one of three possible stop codons.