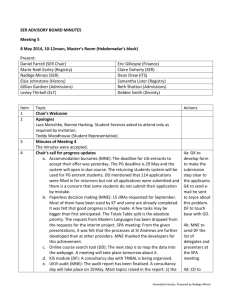
prot24621-sup-0001-suppinfo01
advertisement
Supplementary Material Predicting the side-chain dihedral angle distributions of non-polar, aromatic, and polar amino acids using hard sphere models Alice Qinhua Zhoua,b,c, Corey S. O’Hernb,d, Lynne Regana,b, * aDepartment of Molecular Biophysics & Biochemistry, Yale University, New Haven, CT, USA bIntegrated Graduate Program in Physical and Engineering Biology (IGPPEB), Yale University, New Haven, CT, USA cHoward Hughes Medical Institute International Research Fellow dDepartments of Mechanical Engineering & Materials Science, Applied Physics, and Physics, Graduate Program in Computational Biology and Bioinformatics, Yale University, New Haven, CT, USA *Corresponding author, Lynne.Regan@yale.edu Table S1: Numbers of dipeptides and α-helical segments in the 1.7Å and 1.0Å Dunbrack databases. “Helix” and “Sheet” refer to structures with φ and ψ angles within ±10◦ of the canonical α-helix (φ = −57◦, ψ = −47◦) and β-sheet (φ = −119◦, ψ = 113◦) values, respectively. † Same as Ref. [1], where all dipeptides are extracted with φ and ψ angles changed to canonical α-helix (φ = −57◦, ψ = −47◦) or β-sheet (φ = −119◦, ψ = 113◦) values. [1] Zhou AQ, Caballero D, O’Hern CS, Regan L. New insights into the interdependence between amino acid stereochemistry and protein structure. Biophys. J. 2013;105:2403– 2411. Table S2: Definitions of the backbone and side-chain dihedral angles. All of the dihedral angles listed range from 0◦ to 360◦, except: 1) χ for the aromatic residues (Phe, Tyr, 2 and Trp), which range from 0◦ to 180◦, and 2) φ and ψ, which range from −180◦ to 180◦. ◦ a shows datasfor Sedata r with backbone dral angle andangle χ1 =s and 60 . χWe show hows forhe Slical er with helical dihe backbone dihes dral 1 = 60 . We sho stick repress etick ntation ofsethisconformation in Columnin 2 Column and thepe rcent of thenumbe r repre ntation of thisconformation 2 and thepe rcent of thenum of Ser dipeptide tructure s (with variation bond length andlebond angle combinaof Sersdipe ptide structure s (within variation in bond ngth and bond angle combi tions) that tions) allow this in Columnin 3.Column In thefourth column, wes how the that conformation allow this conformation 3. In thefourth column, weshow i+ 1 1 i+1 1 Ser Helixdistributiondis oftribution distances Oγi+–H on Ni+ 1n ) atom . offor disO tance Oγ –N and(hydroge Oγ –Hi+ 1n(hydroge on Ni +pairs ) atom pa γ –Ns forand χ =60° The red line indicates the hard 50% sphere limit (σij ), and th 1 66 d distances. Near 50%of Ser in hel red indicate 66s disallowe In the sarea econd row, in we show the limiting clash for Ser i The red lineThe indicat t he hard sphere limitsphe (σi j re ), and t he shaded red es line indicate s the hard limitt hus (σij ), and thus the areapale shaded in pale Ser Sheet Oγ –O only 7%areallowe d. Swe imilarly, a comparison o red indicat es ances.dNear 50%s.ofNe Ser helical allowed.areallo redisallowed d indicatesdist disallowe distance ar in 50%of Sebackbone rand in he licalare backbone d. χ =60° 1 tion with on he a ehe lical In t he second row, show ingthe clash for Ser inhβ-sheet conformat is conformation 7%is (Row 3) shows t In the sewe cond row,t he welimit show limiting clas for SerCys in β-s tion conformation is saeve limiting – only 5% is allowed due to Sγ – Oγ –O and only 7%and areonly allowed. Similarly,d.a S comparison of more Ser in on helical Oγ –O 7%areallowe imilarly, a comparis ofre Sly er conformain a helical conforma- t ion red wit hline Cys onre adhelical conformat (Row t hat t hows hearea larger atpale om tion with Cys on a he conformation (Row 3)t he s the large r sulfur atom oxyge nthat in sulfur S er. in Finally, we compare Row 3 and Row The indicat es t he hard sphere limit (σi3) ),shows and tthan hus shaded j re The line indicate slical theion hard sphe limit (σ ij ), and thus the area shaded in pale i+ 1 i + i1+ 1 clashes, i+ 1 Cys Helix is severely limit –lyonly 5% is– allowed due t in o Shelical –Ndue and is seing vesre limiting only is allowe d S –N –Hareallo heve s, ry low percent of bo redmore indicat es dist ances. 50%5% Ser allowed. γmation, γ –H γS χare =and 60◦Sγor 180◦clas . A redisallowed dmore indicate dis allowe dNear dis tance s.ofNe ar 50%of Sebackbone rto inwith he lical we d. 1 backbone χ =60° 1 oxyge ◦ t han oxygen Ser. Finally, we compare Row 3 and Row CysS in helical conforthan nshow inrow, Se r. Finally, wecompareRo w 3 and Row Cys in a he conforIn t he second row, t he limit ing clash for Ser in4: islicalwith 5% Ininthe sewe cond we show the limiting clas hβ-sheet for er aconformat in4: β-s he e tion conformation combinations are compatible χis 1 = 60 , whereas virt ◦ ◦ mation, with =◦ .60 180 .S A very low pe nt of bond leconformangth and angle mat ion, witonly h γχ–O =and 60 orχ180 A◦ or very low percent ofabond and angle 17%areallowe Oγ –O and are allowed. Similarly, comparison of rce Serlengt ahhelical 17% O only d.a imilarly, comparis on of S◦erbond in a he licalbond conformawith χin 1 = 180 . ◦ ◦ combinations are compatible with χ3) 60 , whe virtually 100% are are compat withe hlical χ 1 ion =conformation 60 , whereas virt 100% aresulfur compat 1= tcombinat ion wit hions Cys onwith a helical conformat (Row shows tually hat tas he larger atible om tion Cysible on a (Row 3)re s hows that the large r scompatible ulfur atom 5.2.3 Val and Thr ◦ ◦Cys i+ 1 i + i1+ 1 clashes, i + 1 with χ1Helix =eing 180 witmore h χ 1 severely = 180 . limit is 5% is– allowed t o Sγ d –Ndue andSγS–N is more s ve re–ly.only limiting only 5%due is allowe to Sγ –H clashes, γ –H and 5.2.4 Le u and Ile χ =180° 1 oxyge t han oxygenthan in Ser. Finally, n in Se we r. compare Finally, wecompareRo Row 3 and Row w 4: 3 and CysRow in a 4: helical Cys in confora helical confor98% 5.2.3 5.2.3 Val and ThrVal and Thr ◦ ◦ with =◦ .60 180 . A very low percelengt nt ofhbond lengthangle and bond angle mat wit mation, h χ 1Ile = Le 60 orχ180 A◦ or very low percent of bond and bond 1Ile 5.2.4 u and 5.2.4ion,Leu and ◦ ◦ combinations are compatible with χ1 = 60virt , whe reas virtually 100%ible are compatible combinat ions are compat ible wit h χ 1 = 60 , whereas ually 100% are compat ◦ χ of the steric clashes in Ser and Cys dipeptide mimetics. with 180◦ . wit h χ 1 = Figure 180 . S1: 1 =Illustration Row 1: Ser in an α-helical backbone conformation with side-chain dihedral angle χ1=60◦. 5.2.3 Val inand Thr 5.2.3 ValRow and Thr 2: Ser a β-sheet backbone conformation with side-chain dihedral angle χ =60◦; 1 an Ile α-helical backbone conformation with side-chain dihedral angle 5.2.4 Leuinand 5.2.4 LeuRow and 3: IleCys ◦ χ =60 ; Row 4: Cys in an α-helical backbone conformation with side-chain dihedral 1 angle χ1=180◦. Column 1: Specified backbone conformation and χ1 value in each row. Column 2: Stick representation of Ser and Cys dipeptide mimetics in backbone and side-chain conformations specified in Column 1. Column 3: The separation distributions between key atom pairs (in Å). The red vertical line indicates contact between the pair of atoms. The area shaded in pale red highlights sterically disallowed atomic separations. Column 4: The percentage of Ser residues in the 1.7Å database for which the highlighted atom pairs in Column 3 possess separations that are sterically allowed in the specified conformation (Column 1). 67 67 67 Figure S2: Error bars for the side-chain dihedral angle distributions for Ser and Cys dipeptide mimetics: Comparison of the observed (red lines) and calculated (blue lines) probability distributions P(χ1) of the side-chain dihedral angle χ1 for Ser and Cys in dipeptide mimetics with backbone dihedral angles φ and ψ within ±10° of canonical αhelix (φ=−57°, ψ=−47°) and β-sheet (φ=−119°, ψ=113°) values. To estimate the error bars, we break the set of structures for a given residue type into groups, where from each group we can obtain a reasonably smooth distribution Pi(χ1), where i=1,...,Ng and Ng is the number of groups. The numbers of structures in each group of observed and calculated for Cys and Ser are 50 and 10, respectively. The average P(χ1) is defined as 𝑁𝑔 1 𝑃(𝜒1 ) = ∑𝑖=1 𝑃𝑖 (𝜒1 ) with error bars given by the error in the mean σ(χ1)/(Ng)1/2, where 𝑁𝑔 σ(χ1) is the standard deviation in each bin. The probabilities are normalized such that ∫ P(χ1) dχ1=1. c h i1 0 .1 0 .0 5 c h i2 0 0 .2 0 .1 c h i3 0 0 .0 3 0 .0 2 0 .0 1 0 0 6 0 1 2 0 1 8 0 2 4 0 3 0 0 3 6 0 Figure R1: (left) Stick representation of the Met dipeptide. (right) Predicted probability distributions for χ1 , χ2, and χ3 from the hard-sphere plus stereochemical constraint model for Met dipeptides with two hydrogen atoms artificially placed in an sp3 configuration on the sulfur atom. The probabilities are normalized such that ∫ P(χn) dχn=1.