Specht`s Goodness of Fit Estimate and Test for Path Analyses
advertisement
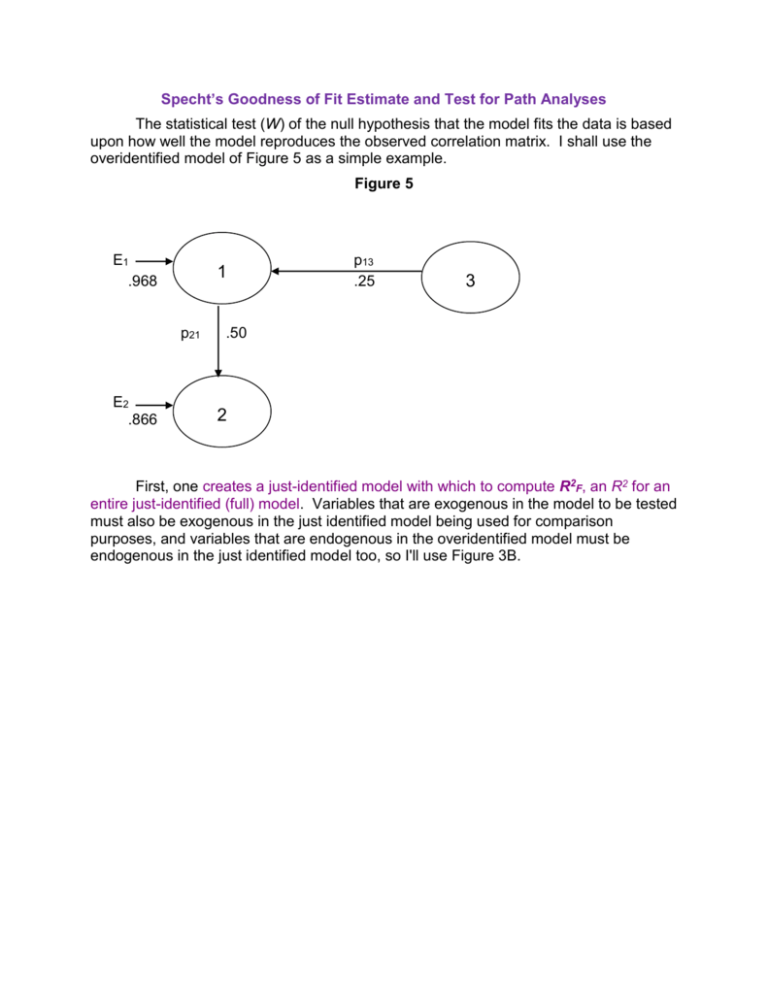
Specht’s Goodness of Fit Estimate and Test for Path Analyses The statistical test (W) of the null hypothesis that the model fits the data is based upon how well the model reproduces the observed correlation matrix. I shall use the overidentified model of Figure 5 as a simple example. Figure 5 E1 .968 1 p21 E2 .866 p13 .25 3 .50 2 First, one creates a just-identified model with which to compute R2F, an R2 for an entire just-identified (full) model. Variables that are exogenous in the model to be tested must also be exogenous in the just identified model being used for comparison purposes, and variables that are endogenous in the overidentified model must be endogenous in the just identified model too, so I'll use Figure 3B. To obtain R2F one simply obtains the product of the squares of all the error paths and subtracts from one. For Figure 3B, R2F = 1 - (.968)2(.775)2 = .437. Next, one computes R2R , an R2 for the overidentified (reduced) model being tested. Again, one simply obtains the product of the squared error path coefficients and subtracts from one. For Figure 5, R2R = 1 - (.968)2(.866)2 = .297. The smaller R2R is relative to R2F , the poorer the fit, and the fit here looks poor indeed. Were the overidentified model to perfectly reproduce the original correlation matrix, R2R would equal R2F, and the fit would be perfect. We next compute Q, a measure of the goodness of fit of our overidentified 1 RF2 Q model, relative to a perfectly fitting (just identified) model. . For our 1 RR2 example, Q = (1 -.437)/(1 -.297) = .801. A perfect fit would give a Q of one; less than perfect fits yield Q's less than one. Q can also be computed simply by computing the product of the squares of the error path coefficients for the full model and dividing by the square of the products of the error path coefficients for the reduced model. For our (.968) 2 (.775) 2 model, Q .801 . (.968) 2 (.866) 2 Finally, we compute the test statistic, W = -(N - d) ln(Q) where N = sample size (let us assume N = 100), d = number of overidentifying restrictions (number of paths eliminated from the full model to yield the reduced model), and ln = natural logarithm. For our data, W = -(100 - 1) * ln(.801) = 21.97. W is evaluated with the chi-square distribution on d degrees of freedom. For our W, p <.0001 and we conclude that the model does not fit the data well. If you compute W for Figure 4A or 4B you will obtain W = 0, p = 1.000, indicating perfect fit for those models. The overidentified model (Figure 5) we tested had only one restriction (no p23), so we could have more easily tested it by testing the significance of p23 in Figure 3B. Our overidentified models may, however, have several such restrictions, and the method I just presented allows us simultaneously to test all those restrictions. Consider the overidentified model in Figure 6. Figure 6 SES 1 E3 p31 .410 r12 .3 .912 p43 nACH 3 .420 p42 .503 IQ 2 GPA 4 .710 E4 The just identified model in Figure 1 is an appropriate model for computing R2F. Figure 1 SES 1 p41 = .009 p31 = .398 nACH 3 r12 = .3 p43 = .416 GPA 4 p32 = .041 E3 p42 = .501 E4 IQ 2 In the just-identified model E3 1 R32.12 .911 and E 4 1 R42.123 .710 . For our overidentified model E3 1 R32.1 .912 and E4 1 R42.23 .710 . R2F = 1 - (.911)2(.710)2 = .582. R2R = 1 - (.912)2(.710)2 = .581. Q = (1 - .582)/(1 - .581) = .998. If N = 100, W = -(100 - 2) * ln(.998) = 0.196 on 2 df (we eliminated two paths), p = .91, our overidentified model fits the data well. Let us now evaluate three different overidentified models, all based on the same correlation matrix. Variable V is a measure of the civil rights attitudes of the Voters in 116 congressional districts (N = 116). Variable C is the civil rights attitude of the Congressman in each district. Variable P is a measure of the congressmen's Perceptions of their constituents' attitudes towards civil rights. Variable R is a measure of the congressmen's Roll Call behavior on civil rights. Suppose that three different a priori models have been identified (each based on a different theory), as shown in Figure 8. Figure 8 Just-Identified Model C .867 EC .498 .324 .075 V R .560 .366 .507 .556 ER P .595 EP For the just-identified model, R2F = 1 - (.867)2(.595)2(.507)2 = .9316. Model X C .766 EC .643 .327 V R .613 .510 .738 ER P .675 EP For Model X, R2R = 1 - (.766)2(.675)2(.510)2 = .9305. Model Y C .867 EC .498 .327 V R .613 .643 .510 ER P .766 EP For Model Y, R2R = 1 - (.867)2(.766)2(.510)2 = .8853. Model Z C .867 EC .498 .327 V R .613 .366 .510 .556 ER P .595 EP For Model Z, R2R = 1 -(.867)2(.595)2(.510)2 = .9308. Goodness of fit Qx = (1 - .9316)/(1 - .9305) = .0684/.0695 = .9842. Qy = .0684/(1 - .8853) = .5963. Qz = .0684/(1 - .9308) = .9884. It appears that Models X and Z fit the data better than does Model Y. Wx = -(116 - 2) ln(.9842) = 1.816. On 2 df (two restrictions, no paths from V to C or to R), p = .41. We do not reject the null hypothesis that Model X fits the data. WY = -(116 - 2) ln(.5963) = 58.939, on 2 df, p <.0001. We do reject the null and conclude that Model Y does not fit the data. WZ = -(116 - 1) ln(.9884) = 1.342, on 1 df (only one restriction, no V to R direct path) p = .25. We do not reject the null hypothesis that Model Z fits the data. It seems that Model Y needs some revision, but Models X and Z have passed this test. Sometimes one can test the null hypothesis that one reduced model fits the data as well as does another reduced model. One of the models must be nested within the other -- that is, it can be obtained by eliminating one or more of the paths in the other. For example, Model Y is exactly like Model Z except that the path from V to P has been eliminated in Model Y. Thus, Y is nested within Z. To test the null, we compute W = -(N -d) ln[(1 - M1)/(1 - M2)] . M2 is for the model that is nested within the other, and d is the number of paths eliminated from the other model to obtain the nested model. For Z versus Y, on d = 1 df, W = -(116 - 1) * ln[(1 - .9308)/(1 - .8853) = 58.112, p < .0001. We reject the null hypothesis. Removing the path from V to P significantly reduced the fit between the model and the data. Karl L. Wuensch Department of Psychology East Carolina University Greenville, NC 27858