jane12442-sup-0001-Supinfo
advertisement
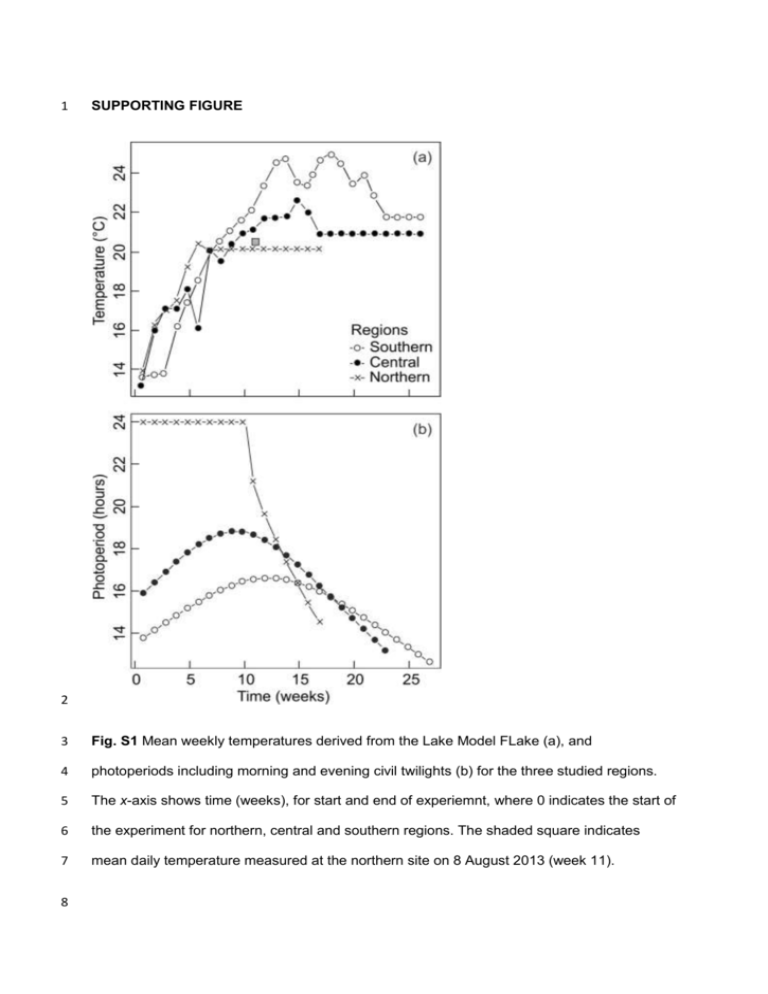
1 SUPPORTING FIGURE 2 3 Fig. S1 Mean weekly temperatures derived from the Lake Model FLake (a), and 4 photoperiods including morning and evening civil twilights (b) for the three studied regions. 5 The x-axis shows time (weeks), for start and end of experiemnt, where 0 indicates the start of 6 the experiment for northern, central and southern regions. The shaded square indicates 7 mean daily temperature measured at the northern site on 8 August 2013 (week 11). 8 9 SUPPORTING TABLES 10 Table S1 11 Coordinates for sampled populations, estimated population size, number of days within a year when the shallow water temperature exceeds 12 10°C, mean shallow water temperature within a growth season, number of degree days for each of the studied populations. Degree days were 13 calculated by multiplying number of days by mean daily temperature during the time when shallow water temperature exceeded 10 °C. 14 Temperatures were derived using the FLake model (Lake Model Flake 2009). 15 16 17 Locality Coordinates Estimated population size (flying adults) No. of days when temperature exceeds 10 °C Mean temperature (°C) within a growth season Degree days France 43°29' N, 4°48' E > 1000 254 19.0 4660.6 France 43°31' N, 4°46' E > 1000 254 19.0 4660.6 Poland 53°29' N, 16°30' E > 1000 175 16.6 2898.7 Poland 53°38' N, 16°22' E 300-1000 175 16.6 2898.7 Sweden 65°36' N, 22°7' E 300-1000 112 15.4 1722.0 Sweden 66°36' N, 19°52' E 50-100 112 15.0 1578.7 18 Table S2 19 Mean (±1 SE) values of egg volume, egg development time, larval development time, larval head width and larval growth rate across studied 20 populations. Locality Coordinates Egg volume (mm3) Egg development time (days) Larval development time (days) Larval head width (mm) Larval growth rate (mm/day) France 43°29' N, 4°48' E 0.070 ±0.0003 35.668 ±0.8168 88.983 ±0.9144 3.594 ±0.0060 0.041 ±0.0004 France 43°31' N, 4°46' E 0.069 ±0.0004 46.633 ±1.4594 86.087 ±1.2103 3.578 ±0.0098 0.042 ±0.0006 Poland 53°29' N, 16°30' E 0.062 ±0.0003 30.762 ±0.5942 86.785 ±0.6594 3.485 ±0.0067 0.041 ±0.0003 Poland 53°38' N, 16°22' E 0.064 ±0.0003 33.765 ±0.6717 85.334 ±0.882 3.493 ±0.0085 0.041 ±0.0004 Sweden 65°36' N, 22°7' E 0.079 ±0.0003 9.691 ±0.0996 78.944 ±0.6269 3.330 ±0.0076 0.043 ±0.0003 Sweden 66°36' N, 19°52' E 0.081 ±0.0005 9.862 ±0.2491 76.107 ±0.7719 3.311 ±0.0102 0.044 ±0.0004 21 Table S3 22 Comparisons of linear mixed models with various constrained (C) and unconstrained (U) 23 structures for respective random effects (S – sire, D – dam, R – residual). Only traits 24 exhibiting significant genetic variance were included. 25 Model no. Trait S D R logLik Models comp. P 26 1.1 Growth rate C C C 3436.45 ‒ ‒ 27 1.2 U U U 3456.62 1.2 vs. 1.1 <0.0001 28 1.3 U U C 3438.84 1.2 vs. 1.3 <0.0001 29 1.4 U C U 3455.72 1.2 vs. 1.4 0.41 30 1.5 C C U 3454.85 1.4 vs. 1.5 0.34 31 1.6 U C C 3455.71 1.6 vs. 1.1 <0.0001 1.4 vs. 1.6 0.99 32 C C C ‒3825.13 ‒ ‒ 2.2 U U U ‒3287.07 2.2 vs. 2.1 <0.0001 35 2.3 U U C ‒3802.10 2.2 vs. 2.3 <0.0001 36 2.4 U C U ‒3303.14 2.2 vs. 2.4 <0.0001 37 2.5 C C U ‒3323.02 2.4 vs. 2.5 <0.0001 38 2.6 U C C ‒3303.80 2.6 vs. 2.1 <0.0001 2.4 vs. 2.6 0.98 33 2.1 34 Devel. time (e) 39 40 3.1 41 3.2 Egg volume C C C 6592.33 ‒ ‒ U U U 6617.90 3.2 vs. 3.1 <0.0001 4 42 3.3 U U C 6595.82 3.2 vs. 3.3 <0.0001 43 3.4 U C U 6615.93 3.2 vs. 3.4 0.14 44 3.5 C C U 6614.28 3.4 vs. 3.5 0.20 45 3.6 U C C 6600.18 3.6 vs. 3.1 0.0004 3.4 vs. 3.6 <0.0001 46 C C C -299.47 ‒ ‒ 4.2 U U U -273.22 4.2 vs. 4.1 <0.0001 49 4.3 U U C -296.32 4.2 vs. 4.3 <0.0001 50 4.4 U C U -273.77 4.2 vs. 4.4 0.58 51 4.5 C C U -274.51 4.4 vs. 4.5 0.63 52 4.6 U C C -297.26 4.6 vs. 4.1 0.11 4.4 vs. 4.6 <0.0001 47 4.1 48 Devel. time (l) 53 54 C C C -40.06 ‒ ‒ 5.2 U U U -37.22 5.2 vs. 5.1 0.22 57 5.3 U U C -38.08 5.2 vs. 5.3 0.42 58 5.4 U C U -39.28 5.2 vs. 5.4 0.12 59 5.5 C C U -39.28 5.4 vs. 5.5 0.39 60 5.6 U C C -40.05 5.6 vs. 5.1 0.98 5.4 vs. 5.6 0.46 55 5.1 56 61 Head width 62 5 63 Table S4 64 Comparisons of linear mixed models with various constrained (C) and unconstrained (U) 65 structures for respective random effects (D – dam, R – residual). Only traits exhibiting 66 significant genetic variance were included. 67 Model no. Trait D R logLik Models comp. P 68 1.1 Growth rate C C 3806.91 ‒ ‒ 69 1.2 U U 3830.01 1.2 vs. 1.1 <0.0001 70 1.3 U C 3807.95 1.2 vs. 1.3 <0.0001 71 1.4 C U 3829.83 1.2 vs. 1.4 0.84 1.4 vs. 1.1 <0.0001 72 C C ‒4231.66 ‒ ‒ 2.2 U U ‒3615.17 2.2 vs. 2.1 <0.0001 75 2.3 U C ‒4194.71 2.2 vs. 2.3 <0.0001 76 2.4 C U ‒3665.85 2.2 vs. 2.4 <0.0001 2.4 vs. 2.1 <0.0001 73 2.1 74 Devel. time (e) 77 C C 7322.98 ‒ ‒ 3.2 U U 7357.62 3.2 vs. 3.1 <0.0001 80 3.3 U C 7324.85 3.2 vs. 3.3 <0.0001 81 3.4 C U 7356.01 3.2 vs. 3.4 0.14 78 3.1 79 82 Egg volume 3.4 vs. 3.1 83 6 C C -2202.30 ‒ 4.2 U U -2169.11 4.2 vs. 4.1 <0.001 86 4.3 U C -2199.18 4.2 vs. 4.3 <0.001 87 4.4 C U -2169.37 4.2 vs. 4.4 0.77 4.4 vs. 4.1 <0.001 ‒ 84 4.1 85 Devel. time (a) 88 ‒ C C 1897.65 ‒ 5.2 U U 1899.10 5.2 vs. 5.1 0.58 91 5.3 U C 1898.13 5.2 vs. 5.3 0.38 92 5.4 C U 1898.50 5.2 vs. 5.4 0.55 5.4 vs. 5.1 0.43 89 5.1 90 93 Head width 94 7 95 APPENDIX 1 96 Study species 97 The damselfly L. sponsa is characterized by a range distribution across central and northern 98 Europe. Being obligatorily univoltine (one generation per year), it is seasonally time-stressed 99 in temperate regions, where it must complete larval development, emerge and mate before 100 the season ends (Dijkstra 2006b; Śniegula & Johansson 2010). Females insert their eggs 101 into plant tissues and the eggs enter winter diapause about two weeks later (Corbet 1956a). 102 Eggs overwinter either above (in plant stems) or below (lake bottom) water surfaces. Winter 103 diapause terminates during winter and post-overwintering egg development (hereafter called 104 egg development) proceeds during the following spring. Larval hatching starts when the 105 water temperature reaches 10°C (Corbet 1956a; Van Doorslaer & Stoks 2005). Spring egg 106 development is, to a great extent, regulated by photoperiod (Śniegula & Johansson 2010). 107 Larval development time is dependent on temperature and photoperiod and takes about two 108 to three months (Corbet 1956b; Pickup, Thompson & Lawton 1984; Pickup & Thompson 109 1984; Śniegula & Johansson 2010; Johansson, Śniegula & Brodin 2010). Although there is 110 no data available on L. sponsa, studies on damselflies in general indicate that the proportion 111 of a female’s offspring that is sired by the last male with which she copulated rarely falls 112 below 95% (Corbet 1999). However, it has been shown that in some species the proportion 113 may vary from 44% to over 90% (Fincke 1984; Cooper, Miller & Holland 1996). 114 115 Field sampling 116 To estimate growth and development in the egg and larval stages, we collected eggs from 117 adult females in three geographic regions covering a distance of 2,730 km: northern Sweden 118 (66°N, alt. 220 mamsl and10 mamsl), north-western Poland (54°N, alt. 140 mamsl) and 119 southern France (43°N, alt. 0 mamsl), hereafter northern, central and southern populations or 120 regions. Estimated number of flying individuals at each sampling site is presented in Table 8 121 S1. One of the northern population was situated beyond the species’ northern geographic 122 distribution shown in Dijkstra (Dijkstra 2006a) and hence can be regarded as a peripheral 123 population. The number of flying individuals during the peak of the flying season (Table S1) 124 as well as our visits in previous years (Śniegula et al. 2014; unpublished data) suggests that 125 the peripheral population is at least several years old. The southern populations are not 126 peripheral, since the species distribution extends further south. We sampled two populations 127 within each study region. The distance between northern sites was 151 km. The central 128 populations were separated by 18 km, the southern by 5 km. We sampled two populations in 129 each study region to consider possible intraregional, cross-population variation in study traits. 130 Our results show that the two sampled populations in northern, central and southern 131 populations did not differ in genetic variance. One sampling site in the northern, two in central 132 and two in southern regions supported fish populations. We have no information on the 133 status of fish in the second site in northern region. Field sampling started with the southern 134 and ended with the northern populations (southern population 29 June and 2 July, central 135 23‒28 July, and northern 6‒10 August). We collected paternal half-sibling egg clutches from 136 the one northern, two central and two southern populations. This enabled us to estimate 137 additive genetic variance, taking into account potential maternal effects (Lynch & Walsh 138 1998). We collected these paternal half-sibs by separating the initially copulating pairs and 139 saving females for egg laying. The male mating with the first female was thereafter enclosed 140 in a small insectary together with a new single female. Using this method we produced the 141 following number of paternal half-sib families: northern site, 10 males, each of which mated 142 with two females, resulting in a total of 20 families; central sites, 16 and four males, each of 143 which mated with two females, resulting in a total of 32 and eight families, respectively; and 144 southern sites, 18 and nine males, each of which mated with two females, resulting in a total 145 of 36 and 18 families, respectively. In addition, from the populations where we sampled full- 146 sibs, the northern, one central and one southern population we sampled the following 147 numbers of adult females that produced full-sib families: eight, nine and one respectively. 148 These families were also included in the variance partitioning analysis (see below). The less 9 149 numerous northern population did not allow us to collect half sibs. However, we obtained 16 150 full-sib families from this northern population. 151 After copulation, females were individually placed in plastic jars with moist filter 152 papers as oviposition substrates and transported to ‘research stations’ situated up to 16 km 153 away from the sampling sites. These females were kept in room conditions (temperature 154 22°C) by a window, i.e. with a natural photoperiod at the sampling site. We kept females in 155 this condition until they oviposited eggs, an event which occurred within 48 hours after field 156 sampling. Egg clutches were stored in these conditions for up to five days (i.e. until all 157 females laid eggs). Then we placed the clutches, embedded in filter paper, in dark conditions 158 (Styrofoam boxes with an interior temperature of 22°C) and transported them by car to the 159 laboratory (Cracow, Poland). Transport of egg clutches took from one (central populations) to 160 three (northern populations) days. Our previous experiments indicated that such transport 161 has little or no effect on damselfly development (Śniegula & Johansson 2010; Śniegula et al. 162 2014); therefore we feel confident that this method excludes unwanted effects on life history 163 traits. 164 165 Experimental set-up 166 Estimates of growth and development of eggs and larvae were carried out at the Institute of 167 Nature Conservation PAS in Cracow, Poland, in climate chambers. In three separate 168 chambers, we reared the northern, central and southern populations at programmed 169 temperatures and photoperiods (thermo-photoperiods) simulating the thermo-photoperiods 170 experienced by the damselflies in their natural conditions (experiment 1, Fig. S1). A fourth 171 chamber, with a mean thermo-photoperiod averaged over all sampled regions and growth 172 seasons, was used to rear all four study regions (experiment 2). Upon the arrival of eggs at 173 the laboratory, the clutches were placed in plastic containers (12x8 cm, 5 cm high) filled with 174 250 ml of mixed dechlorinated tap water and filtered pond water. We thereafter simulated the 175 progress of summer, winter and spring conditions as described below. 10 176 Experiment 1 177 In this experiment we determined genetic variance in life history traits simulating natural 178 temperature and photoperiod regimes by using a half-sib design based on the northern, 179 central and southern populations. One of the northern population was excluded from this 180 analysis, since we could not produce half-sib families. However, it is included in summary 181 statistics and experiment 2. Containers with eggs from each of the study regions were put 182 into three climate chambers with programmed temperatures and photoperiods (thermo- 183 photoperiods) mimicking natural changes in these variables in natural conditions in each 184 sampling region. Two separate models for calculation of water temperature at the two 185 northern sites showed statistically identical mean weekly temperatures during the growth 186 season for L. sponsa larvae (results not shown). To follow the natural progress of the 187 thermo-photoperiod through the season, we changed it every Friday (Fig. S1). The exception 188 was the period of winter simulation (described below). 189 To apply a realistic surface water temperature for each region, we used an extension of 190 the lake model FLake (Lake Model Flake 2009) constructed by (Nilsson-Örtman et al. 2012) 191 to calculate temperatures throughout the season at each sampling location. We thereafter 192 used the mean weekly values for the populations at each region as our temperature 193 treatment (Fig. S1a). The following parameters were included in the model: surface thermal 194 radiation, solar radiation, wind speed, dew point temperature, and air temperature. However, 195 the model did not take temperature inversion, which becomes less frequent at progressively 196 higher temperate latitudes, into account. This can strongly ameliorate harsh environmental 197 conditions, especially near the bottom of the shallow, dark bottom bodies of water that are 198 frequent in boreal regions (Corbet 2003). The northern individuals originated from such 199 bodies of water. We found that water temperatures in the northern region were 1‒2 degrees 200 higher than that derived from the model, supporting stronger temperature inversion in the 201 north. We therefore increased the mean weekly temperatures in chambers holding the 202 northern and central individuals by 2°C (Fig. S1a). 11 203 The applied photoperiodic regimes used for each latitudinal region included morning 204 and evening civil twilights (Fig. S1b). Insects, including odonates, are known to be very 205 sensitive to light intensity and the threshold at which they register light is very low (Lutz & 206 Jenner 1964; Saunders 2002). Note that in natural conditions populations from the northern 207 regions experience the same photoperiods during the larval growth season, i.e. from the end 208 of May to mid-July (24 hours of daylight, including civil twilight). These temperatures and 209 photoperiods were initiated once larvae had overwintered in the laboratory (see below). 210 Upon arrival at the laboratory, the northern, central, and southern eggs were given the 211 following hours of light-dark conditions ‒ L 20:57 and D 3:03, L 17:38 and D 6:22, L 16:31 212 and D 7:29, respectively ‒ along with corresponding temperatures ‒ 19.2°C, 21°C, and 213 24.8°C, respectively ‒ simulating late summer conditions. After three weeks, when eggs had 214 begun diapause (Corbet 1956a), we initiated winter conditions by lowering the temperature to 215 15°C. The next day we changed the temperature to 5°C and switched off the light. This 216 procedure was applied to the northern and central populations. For the southern individuals 217 we used only two weeks of late summer conditions, because some eggs from several 218 clutches started to hatch nearing the end of the second week of their development. The 219 northern, central, and southern eggs were kept dark at 5°C, simulating winter conditions for 220 28±1.5, 28±1, 28±1.5, and 28±1.5 days, respectively (the variation is due to differences 221 between females in ovipositing dates). 222 To simulate spring conditions we set thermo-photoperiods to match the dates when 223 temperatures exceeded 12°C at the origin of the populations. For the northern regions this 224 corresponded to 30 May (temp. 14°C), for the central region 25 April (temp. 13.3°C), and for 225 the southern region 4 April (temp. 13.8°C) (Fig. S1a). These temperatures correspond to the 226 first week after 12°C was reached. We chose these temperatures (and hence photoperiods) 227 for the initiation of spring, since egg hatching in L. sponsa starts when the water temperature 228 exceeds 10°C (Corbet 1956a). Thereafter, we simulated natural (weekly) changes in 229 temperature until 25 July in the chamber housing the northern populations (week 9), 15 12 230 August in the chamber housing the central populations (week 17) and 12 September in the 231 chamber housing the southern populations (week 24). On these dates, when the temperature 232 in nature starts to slowly decline, there were still individuals that had not emerged. We 233 therefore maintained the temperature that the larvae experience at these dates until all 234 individuals had emerged in the northern populations (Fig. S1a). The photoperiod followed 235 weekly changes until the end of the experiment (Fig. S1b). We terminated the experiment on 236 7 February 2014, which corresponded to 17 Oct (northern regions), 26 Sept (central region) 237 and 3 Sept 2014 (southern region) under natural conditions. None of the northern larvae 238 remained on this date (all had emerged), whereas nine central and eight southern outliers 239 were still in the larval stage. We terminated the experiment because from these dates 240 forward the climatic conditions are unlikely to be beneficial for emergence, although it is 241 common to record aged flying individuals at this date in central Europe (Wendzonka 2005). 242 Eggs started to hatch soon after spring conditions were initiated. When this happened, 243 larvae from each family were individually placed in round plastic containers (diameter 7 cm, 244 height 4 cm) and fed daily with a mean of 350 SE: 26.8 laboratory-reared brine shrimp, 245 Artemia salina. Ten larvae were taken from each female, resulting in 440 northern, 490 246 central and 550 southern individuals, for a total of 1,480 individuals at the start of the 247 experiment. 248 We estimated the following life history traits: egg volume, egg development time, larval 249 development time, larval size at last instar (F0) and larval growth rate. After the spring 250 initiation, we photographed 10 randomly chosen eggs from each clutch with a Moticam 3MP 251 digital camera mounted on a Motic SMZ168 microscope. These ten eggs correspond to the 252 ten larvae used for the larval growth experiment. We estimated ellipsoid egg shape volume 253 with the image analysis program Motic Image Plus 2.0 by using the equation V=1/6 x 3.14 x 254 L x W 2, where L is egg length and W is egg width. We calculated egg development time as 255 the number of days from the initiation of spring conditions until hatching. When the larvae 256 reached the last instar, we photographed the larvae to estimate their size. We calculated 13 257 larval size as head width, using the ImageJ v.1.36b image analysis program (see Śniegula 258 2012a, b for details). The larval development time was estimated as the number of days 259 between hatching and emergence. The larval growth rate was estimated as final instar larval 260 head width divided by the number of days needed for larval development, i.e. between 261 hatching and emergence dates. We used head widths for growth rate estimates as this 262 measure significantly correlates with other body size traits and is commonly used for adult 263 size and growth rate estimates (Corbet 1999). In addition, using head width instead of weight 264 at emergence allowed us to use a larger sample size for growth rate, since it was impossible 265 to accurately determine the dry weight of some emerging individuals. 266 14 267 Statistical methods, experiment 1 268 We employed a full-sib/half-sib design, where each sire was mated with two dams and 269 offspring were measured in each full-sib family. Thus, full sibs for each dam were also 270 paternal half sibs. In such a breeding design, the covariance between paternal half sibs 271 (PHS) is equal to the variance between sires (V(s)) and approximates one-quarter of the total 272 additive genetic variance V(a) (Lynch & Walsh 1998). Observed variance between dams V(d) 273 is the sum of several components: ¼ additive genetic variance, ¼ dominance variance, plus 274 several terms related to epistatic effects and maternal effects if present (both genetic and 275 environmental). The remaining sources of variation (e.g. environmental) form the 276 unexplained residual component of the variance (V(e)). Heritability can thus be approximated 277 as 4tPHS, where the intra-class correlation between paternal half sibs (tPHS) is defined as 278 V(s)/V(z), i.e. the fraction of total phenotypic variance V(z)=V(s)+V(d)+V(e) explained by sire 279 effects (V(s)). 280 Data was analyzed using the linear mixed model in ASReml-R v. 3.0 (Butler et al. 281 2009) and the R computing environment (R Development Core Team 2012). Prior to analysis 282 all response variables were standardized (to mean = 0 and SD = 1). In all analyses we 283 inspected residual plots to ensure that the models fitted the data correctly. In all models we 284 included sire and dam identity as random effects, and the region of sampling as a fixed 285 effect. Preliminary analyses, including population identities, indicated no population-related 286 differences in estimated parameters within regions. We thus decided to remove the 287 population effect from all models to increase the power of comparisons. We included 288 offspring sex (male/female/unknown) as a fixed variable; however, we later removed it, as it 289 proved insignificant. In total we analyzed five response variables: egg volume, embryonic 290 developmental time, larval developmental time, last instar (F0) larval head width, and larval 291 growth rate. 292 293 To test for the presence of genetic variance and its partitioning among regions, we employed a hierarchy of mixed models of successively greater complexity. A detailed 15 294 description of the testing procedure and results can be found in Supporting Information 2, but 295 in short, we relaxed constraints placed on the covariance matrices and fitted all random 296 effects as square 3×3 covariance matrices. Testing of respective variances and their 297 differences was performed using the likelihood-ratio test. 298 In half-sib/full-sib cases heritabilities (h2) were calculated as 4(V(s)/V(z)), except in 299 cases where the between-sires and between-dams variances were approximately equal. In 300 the latter case heritability was calculated as 4((V(s)+V(d))/V(z)) (i.e. using the covariance 301 between full sibs as the proxy of ¼Va; Lynch & Walsh 1998). We also calculated fractions of 302 total variance explained by the dam effect m2 = V(d)/V(z). Standard errors of all variance 303 functions were calculated using the delta method (Lynch & Walsh 1998). All values of h2 and 304 m2 were calculated from models with the highest likelihood. 305 Due to logistic constraints it was impossible to gather data on half sibs in the one of 306 the two northern populations. As all these individuals were full sibs it was impossible to 307 separately estimate dam and sire components of variance with these individuals included 308 (these two components are fully confounded in full sibs). Hence, data on individuals from this 309 population were included in summary statistics, overall phenotypic variance analysis and full- 310 sib analyses, but not in the half-sib variance analysis described above. In the analysis based 311 on full-sib design, we have treated the two northern populations as coming from one region. 312 The structure of random effects in these models was different as it did not include sire effect. 313 Broad-sense genetic variance was approximated in those models by the dam (i.e. family) 314 effect. In purely full-sib analyses broad-sense heritabilities for all traits were calculated as 315 2*V(d)/V(z) (Lynch & Walsh 1998). 316 To estimate regional phenotypic differences in egg volume, embryonic and larval 317 development time, F0 larval size and larval growth rate (larval size/larval development time in 318 days), we used a linear mixed model function implemented in the package for R nlme, where 319 full-sib families and populations were random effects. 320 16 321 Experiment 2 322 In this experiment we determined genetic variance in life history traits using a constant 323 temperature and photoperiod and a full-sib design. We used this design because space 324 limitation in the climate chamber did not allow for a half-sib design with enough replicates; 325 hence half sibs were not included. The main aim was to compare the difference in genetic 326 variance expressed in non-native, average conditions and at the simulated natural 327 temperature and photoperiod used in experiment 1. When the winter simulation in 328 experiment 1 was terminated (see above), we randomly chose six eggs from eight randomly 329 chosen full-sib families from northern, central and southern regions. This resulted in a total of 330 144 larvae, which were then placed in a chamber with a constant temperature of 21.9°C and 331 a photoperiod corresponding to the longest day length during the growth season (summer 332 solstice, June 21) at a mid-latitude along the transect of our study regions (55°N, 10°E), L 333 19:25, D 04:35. We set this temperature because (1) earlier studies indicated that larvae are 334 characterized by the lowest mortality when reared at this temperature (Johansson et al. 335 2001; Stoks, De Block & McPeek 2006; Śniegula & Johansson 2010) and (2) this 336 temperature is experienced by all study regions in natural conditions for at least several 337 hours within a day during the peak in the growth season. We used a constant temperature 338 and photoperiod because we wished to estimate whether the amount of genetic variance in 339 the studied traits changed as the individuals were grown in a constant and changing native 340 temperature and photoperiod. In this experiment we estimated the same life history 341 parameters as in experiment 1, except we did not measure egg volume. 342 343 Statistical methods, experiment 2 344 In this experiment we used a full-sib design. All sampled populations, including the two 345 northern ones, were used for estimations. To test whether family effects (i.e. broad-sense 346 genetic effects, G) are correlated between two contrasting environments (E): simulated 17 347 natural thermo-photoperiods (experiment 1) and a constant mean thermo-photoperiod for all 348 regions (experiment 2), we fitted an additional set of mixed models in which, for each 349 response variable, we included region and experimental group (simulated vs. constant 350 conditions) as a fixed effect. The random family effect was fitted in the form of four different 351 (co)variance structures: 352 1) Homogenous (equal) variances 353 2) Heterogeneous variances and family-wise correlation between treatments equal to unity 354 3) Heterogeneous variances and family-wise correlation between treatments equal to zero 355 4) Heterogeneous variances and family-wise correlation unconstrained 356 All models were fitted in ASReml-R (Butler et al. 2009). Significance of the interaction of 357 genetic effects and conditions (i.e. the presence or absence of genetic correlation between 358 simulated and constant conditions) was tested using a likelihood-ratio test. Comparison of 359 model 1 and 2 tests the presence of G×E interaction due to uneven genetic variances; 360 comparison of models 2-3 and 3-4 tested for G×E due to cross-environmental correlations of 361 genetic effects being less than one. For visualization purposes we extracted BLUPs (best 362 linear unbiased predictors) of the genetic family effect (Robinson 1991) from all best-fitting 363 models. BLUPs were used solely for graphing purposes 18 364 APPENDIX 2 365 Genetic variance and its partitioning among regions 366 To test for the presence of genetic variance and its partitioning among regions, we employed 367 a hierarchy of models of successively greater complexity. To test for the presence of 368 significant genetic variance we have compared the model with the sire, dam and residual 369 effects to the model with the sire effect excluded. Further analyses comprised a combination 370 of various (co)variance structures defined for the three random effects (sire, dam and 371 residual) and structured according to the three studied regions. All models were first fitted 372 assuming a homogenous covariance structure for regions, i.e. assuming equal variances in 373 all random effects in separate regions. We then tested whether the variance could be 374 partitioned with respect to regions – i.e. whether different regions exhibited different genetic 375 variances. In these analyses we relaxed constraints placed on the covariance matrices and 376 fitted all random effects as square 3×3 covariance matrices. The covariances between 377 regions with respect to each random effect were fixed at zero. We compared the following 378 sets of models (constrained (C) – one variance in all three regions; unconstrained (U) – 379 separate heterogeneous variance in three regions): 380 1) all random effects unconstrained vs. all random effects constrained 381 2) all random effects unconstrained vs. residual variance constrained 382 3) all random effects unconstrained vs. dam variance constrained 383 4) all random effects unconstrained vs. dam and sire variance constrained 384 5) sire variance unconstrained vs. all random effects constrained 385 6) sire and residual variance unconstrained vs. sire variance unconstrained 386 Comparisons 1, 5 and 6 directly tested the hypothesis of ‘no differences in sire’ (hence 387 genetic) variance between regions, employing various constraints on the remaining random 19 388 effects. The rationale behind these comparisons stems from the fact that the model allowing 389 for unconstrained (i.e. differing) genetic variances between regions should fit the data worse 390 than the constrained, equal-variances model if genetic variances in different regions are 391 identical. The degree of this lack-of-fit can be measured by the difference of the likelihoods of 392 the two models, asymptotically distributed as a chi-squared variate with degrees of freedom 393 equal to the number of additional parameters varying in the unconstrained model. The 394 remaining comparisons were performed in order to further study heterogeneity in other 395 random effects and possibly account for their confounding influence on differences observed 396 in genetic variance. The applied hierarchy of models (see Table S3) reflects the logical order 397 of testing in search for significant region-specific variances: in the first step we ask whether 398 the variances differ at all, then we verify that this difference stems from the sire effect 399 (holding all the remaining random effects constrained), and finally we test whether residual 400 variance alone is not the confounding factor generating the observed differences in region- 401 specific sire variances. 402 Full-sib data analyses were performed in the same way with one modification: the sire 403 effect was absent in all models and hence only two random effects were tested (dam and 404 residual effect). Consequently, the number of different models to compare is reduced (see 405 Table S4). 406 20 407 REFERENCES 408 409 410 411 Butler, D.G., Cullis, B.R., Gilmour, A.R. & Gogel, B.J. (2009) ASReml-R Reference Manual. VSNI International. Cooper, G., Miller, P.L. & Holland, P.W.H. (1996) Molecular genetic analysis of sperm 412 competition in the damselfly Ischnura elegans (Vander Linden). Proceedings of the 413 Royal Society of London Series B-Biological Sciences, 263, 1343–1349. 414 Corbet, P. (1956a) The influence of temperature on diapause development in the dragonfly 415 Lestes sponsa (Hansemann) (Odonata: Lestidae). Proceedings of the Royal 416 Entomological Society of London. Series A, General Entomology, 31, 45–48. 417 Corbet, P. (1956b) The life-histories of Lestes sponsa (Hansemann) and Sympetrum 418 419 420 421 striolatum (Charpentier)(Odonata). Tijdschrift voor Entomologie, 99, 217–229. Corbet, P. (1999) Dragonflies: Behavior and Ecology of Odonata. Harley Books, Colchester, UK. Corbet, P. (2003) A positive correlation between photoperiod and development rate in 422 summer species of Odonata could help to make emergence date appropriate to latitude: 423 a testable hypothesis. Journal of the Entomological Society of British Columbia, 100, 3– 424 17. 425 426 427 428 Dijkstra, K.-D.B. (2006a) Field Guide to the Dragonflies of Britain and Europe. British Wildlife Publishing, Gillingham. Dijkstra, K.-D.B. (2006b) Field Guide to the Dragonflies of Britain and Europe. British Wildlife Publishing, Gillingham, UK. 429 Van Doorslaer, W. & Stoks, R. (2005) Thermal reaction norms in two Coenagrion damselfly 430 species: contrasting embryonic and larval life-history traits. Freshwater Biology, 50, 431 1982–1990. 432 Fincke, O. (1984) Sperm competition in the damselfly Enallagma hageni Walsh (Odonata, 433 Coenagrionidae) - benefits of multiple mating to males and females. Behavioral Ecology 434 and Sociobiology, 14, 235–240. 21 435 Johansson, F., Śniegula, S. & Brodin, T. (2010) Emergence patterns and latitudinal 436 adaptations in development time of Odonata in north Sweden and Poland. 437 Odonatologica, 39, 97–106. 438 439 440 441 442 443 444 445 446 Johansson, F., Stoks, R., Rowe, L. & De Block, M. (2001) Life history plasticity in a damselfly: effects of combined time and biotic constraints. Ecology, 82, 1857–1869. Lake Model Flake. (2009) FLake online. Available online http://www.flake.igbberlin.de/index.shtml Lutz, P.E. & Jenner, C.E. (1964) Life-history and photoperiodic responses of nymphs of Tetragoneuria cynosura (Say). Biological Bulletin, 127, 304–316. Lynch, M. & Walsh, B. (1998) Genetics and Analysis of Quantitative Traits. Sinauer Associates Inc., Massachusetts, USA. Nilsson-Örtman, V., Stoks, R., De Block, M. & Johansson, F. (2012) Generalists and 447 specialists along a latitudinal transect: patterns of thermal adaptation in six species of 448 damselflies. Ecology, 93, 1340–1352. 449 Pickup, J. & Thompson, D. (1984) The effects of prey density and temperature on 450 development of larvae of the damselfly Lestes sponsa (Hans.) (Zygoptera: Lestidae). 451 Adv. Odonatol., 2, 169–176. 452 453 Pickup, J., Thompson, D. & Lawton, J. (1984) The life history of Lestes sponsa (Hamsemann): Larval growth (Zygoptera: Lestidae). Odonatologica, 13, 451–459. 454 R Development Core Team. (2012) R: A Language and Environment for Statistical 455 Computing. The R Foundation for Statistical Computing, Vienna, Austria. 456 457 Robinson, G.K. (1991) That BLUP is a good thing: The estimation of random effects. Statistical science, 15–32. 458 Saunders, D.S. (2002) Insect Clocks. 3rd Edition. Elsevier. 459 Śniegula, S., Drobniak, S.M., Gołąb, M.J. & Johansson, F. (2014) Photoperiod and variation 460 in life history traits in core and peripheral populations in the damselfly Lestes sponsa. 461 Ecological Entomology, 39, 137–148. 22 462 463 464 465 466 467 Śniegula, S. & Johansson, F. (2010) Photoperiod affects compensating developmental rate across latitudes in the damselfly Lestes sponsa. Ecological Entomology, 35, 149–157. Stoks, R., De Block, M. & McPeek, M. (2006) Physiological costs of compensatory growth in a damselfly. Ecology, 87, 1566–1574. Wendzonka, J. (2005) Identification key to the imagines of Polish dragonflies (Odonata). Odonatrix. Bulletin of the Odonatological Section of the Polish Entomological Society. 468 23