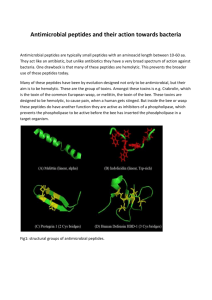
DIPP = Directed Immobilization of Pre
advertisement

Table 1. Bibliography of applications using peptide (micro)arrays for enzyme profiling and substrate identification. The table lists aim of the studies, screening molecules, detection methods, displayed peptide libraries, array preparation technologies, and each publication citation. Entries were arranged according to the enzymatic reaction and within each reaction type according to the year of publication. In the case of kinases entries were divided into subgroups. Aim Screening molecule Detection Library displayed on peptide array Techn ology Ref. PKA, TRI, TRII cytoplasmic domain 32P-ATP and phosphorimaging Combinatorial peptide library SODA (1) PKC, PKA, CK1 and CK2 32P-ATP and phosphorimaging SPOT (2) Kinases: Substrate Identification Determination of substrate specificity Determination of substrate specificity X ≠ Ser, Thr, Cys 23 Mammalian PKC substrate peptides and aa replacements (P, K, L, E, A) Determination of substrate specificity ANPK 32P-ATP and phosphorimaging Kinase substrates known from literature SPOT (3) Substrate specificity determination PKA, PKG 32P-ATP and phosphorimaging Combinatorial peptide library and interative deconvolution SPOT (4) libraries: XXX12XXX, X = mixture of all 20 aa, 1,2 = individual aa Determination of substrate specificity Dyrk1A (R. norvegicus) and truncation variant 32P-ATP and phosphorimaging Dyrk1A substrate peptide RRRFRPASPLRGPPK and 300 aa substitutions SPOT (5) Determination of substrate specificity CDPK-1 (from maize seedlings) 32P-ATP and phosphorimaging Peptide LARLHSVRER, truncations and aa substitutions (L, A, R, E) SPOT (6) Determination of substrate specificity and inhibitor screen PKA and PKG 32P-ATP and phosphorimaging Combinatorial peptide libraries and interative deconvolution SPOT (7) libraries: XXX12XXX, XXXRK12X, XRKKK12X, 1RKKKKK2, LRKKKKKH12, 12LRKKKKKH, 1,2 = defined aa, X = mixture of all aa except Ser and Thr Determination of substrate specificity NEK6 wt and inactive variant 32P-ATP and phosphorimaging 710 peptides from annotated human phosphosites, DIPP (8) DIPP (9) substitutional analysis of GLAKSFGSPNRAY Determination of substrate specificity p60c-Src Fluorescein labelled anti-pTyr ab Combinatorial library, start: YIYGSFK, deletion library, Ala scan, positional scan and combinatorial library XXXXYXXXX, X ≠ Cys, Tyr Profiling of phosphospecific antibodies and detection of kinase substrates CK2 32P-ATP and phosphorimaging or fluorescein labelled anti-pTyr ab (Pt66, Sigma Aldrich), anti- pSer and pThr abs 2304 phosphorylated and non phosphorylated peptides from annotated human phosphosites for ab profiling and 11096 peptides covering the cytoplasmic domains of all human membran proteins and 700 biotinylated peptides for comparison of phosphorylation on microarrays and in solution DIPP (10) Determination of substrate specificity for substrate prediction Abl 32P-ATP and phosphorimaging 1433 Random sequences 13meric peptides with fixed tyrosine residue in the middle position DIPP (11) Determination of substrate specificity M.musculus and H.sapiens TSSK3 32P-ATP and phosphorimaging 200 Peptides DIPP (12) Determination of substrate specificity KPI-2 33P-ATP and phosphorimaging 1152 Kinase substrate peptides of mixed DIPP (13) species Substrate specificity determination and comparison Dyrk1A, Dyrk2, Dyrk4 33P-ATP and phosphorimaging 720 Peptides derived from human phosphosites DIPP (14) Determination of substrate specificity MELK 32P-ATP and phosphorimaging 192 Peptides comprising in vivo phosphorylation sites and peptides containing deletions or aa substitutions DIPP (15) Determination of substrate specificity Dbf1-Mob1 (S. cerevisiae) and inactive variant N305A 32P-ATP and phosphorimaging Oriented peptide library and interative deconvolution libraries, DIPP (16) DCSD (17) library: MAXXXXRXXSXXXXAK KK, proteome array containing 4400 GST proteins from S. cerevisiae and peptide microarray with 2296 annotated human phosphosites Kinase substrate screening Abl, Her2, VEGFR2/KDR Anti-pTyr ab (PY20, Exalpha Biologicals), sec. ab Cy3 labelled 10000 member PNA encoded peptide library: F-Q-AA4-AA3-Y-AA2AA1-Ile-Lys AA1, AA2, AA4: I, V, F, P, R, D-P, S, D-V AA3: I, V, A, P, R, K, -A, S, P Substrate specificity determination MPK1-3 (S. lycopersicum) and MPK6 (A. thaliana) 33P-ATP and phosphorimaging 976 Annotated kinase substrate consensus sites DIPP (18) Determination of substrate specificity Plk4/Sak (M. musculus) wt and inactive catalytic domain 32P-ATP and phosphorimaging DYDIPTTENLYFQ, substitutional analysis of MSYYHHHHHH, substitutional scan of RKKKSFYFKKHHH and KSFYFKKHHH and 32 peptides from proteins containing putative phosphorylation sites and variants of positive peptides SPOT (19) Determination of substrate specificity p60 c-Src, Abl and cytoplasmic domains of EphA4 32P-ATP and phosphorimaging Array containing 144 Tyr kinase substrate peptides, array containing all cytoplasmic Y ontaining motifs from 2chimaerin, claudin-4 and FGFR1 and array containing systematic aa substitutions of Dok6 Y220 and RasGAP Y386 derived peptides and multiple substitutions SPOT (20) Verification of the dual specificity of CK2 CK2, CK2 32P-ATP and phosphorimaging CK2 derived peptides and sequences based on S. cerevisiae Frp3 togehter with positional scan SPOT (21) Substrate specificity determination Protein kinase 7 (P. falciparum) 33P-ATP and phosphorimaging 1176 Kinase substrate peptides DIPP (22) Use of peptide arrays for prediction of kinase substrate specificities c-Raf, MAP3K8 33P-ATP and phosphorimaging 1176 Peptides containing human phosphorylation sites and second array containing 1024 peptides DIPP (23) Substrate identification FLT3, inactive FLT3 K644A PDGFR and inhibitors STI571, AG1269 32P-ATP and X-ray film imaging at –70 °C 384 Peptides derived from known phosphorylation sites DPCC (24) Substrate specificity determination p60c-Src, Abl, EGFR, JNK1, Erk2, p38, PKA Phos-tag-biotin followed by fluorescence labelled streptavidin, Cy5 labelled anti-pTyr ab > 290 Tyr containing peptides, > 1100 Ser or Thr containing peptides and new PKA substrates: RKRRGSLDESD, RKRRASLDESD, RKRRGSFDESD, RKRRASFDESD, TKRSGSVYEPL, GAQAASPAKGE, ALRRApSLGW, ALRRASLGW, ALRRAALGW DIPP (34) Analysis if PknB can phosphorylate human peptides and substrate PknB from S. aureus 33P-ATP and phosphorimaging 976 Disease related human kinase phosphorylation sites DIPP (26) specificity determination Determination of substrate specificity followed by p53 homology screen DAPK, Chk1 33P-ATP and phosphorimaging 192 Kinase peptide substrates DIPP (27) Substrate specificity determination PKA isoforms Tpk1 and Tpk2 from S. cerevisiae 33P-ATP and phosphorimaging SDLRRTS, LRRTSIIGTI, GRQRRLSSLSEF, QQTRRGSEDDTY, LRRASLG and aa substitutions SPOT (28) SPOT (29) Kinases: Identification of phosphorylation sites in substrate proteins Determination of Lyn Anti-pTyr ab (4G10, 23 PKC derived phosphorylation sites Upstate), HRP sec. peptides with central Tyr Ab in PKC residue 32P-ATP and Edman sequencing C-CAM-L derived peptides, Glu variants and Ala screen SPOT (30) Gsk-3 -1 stimulated Hela lysate 32P-ATP and phosphorimaging p65 peptide scan and systematic mutagenesis of Ser, Thr and Tyr to Ala SPOT (31) Detection of phoshorylation site in AKAP-Lbc PKA 32P-ATP and phosphorimaging 20mer peptides encompassing Cterminus of AKAP-Lbc SPOT (32) Determination of CK2 phosphorylation sites in SSRP1 CK2 32P-ATP and phosphorimaging SSRP1 derived peptides containing Ser or Thr and Ala variants SPOT (33) Identification of kinases responsible for identified phosphorylation sites PKA, Akt, PKC, PKC, PKC, CK2, p38, Erk1, Cdk5 32P-ATP and phosphorimaging 190 Peptides encompassing 95 phosphosites from MS analysis and control peptides DIPP (34) Phosphorylation site determination Recombinant human EphA4 receptor tyrosine kinase 33P-ATP and phosphorimaging Overlapping 13meric peptides corresponding to the juxtamembran region of hEphA4 SPOT (35) Phosphorylation site determination Recombinant human EphA4 receptor tyrosine kinase 33P-ATP and phosphorimaging Overlapping 13meric peptides corresponding to the juxtamembran region of hEphA4 SPOT (35) Determination of phosphorylation sites in large cell-cell adhesion molecule (CCAM-L) PKC Detection of phosphorylation sites on p65 (S. scrofa) Identification of the kinase responsible for T840 phosphorylation of GluR1 PKA, Akt1, PKC, PKC, PKC, PDK1, Cdk5, Raf1, p38, MEK1, Erk1, Erk2, Rsk2, JNK3, Gsk3, CaMK2, CK1, CK2, S6K, Rock2, TBK1, p60 c-Src, Fyn, Fes, Pyk2 32P-ATP and phosphorimaging NEAIRTSTLPRNSGA, NEAIRTSVLPRNSGA and peptides containing consensus phosphorylation sites DIPP (36) Validation of new c-Src substrates identified by mass spectrometry p60 c-Src 32P-ATP and phosphorimaging 312 Peptides encompassing all Tyr residues from 14 selected proteins and Phe variants DIPP (37) Phosphorylation site determination CaMK2 32P-ATP and phosphorimaging 23 Selected peptides covering Ets-2 Ser or T residues and positive control pp28 SPOT (38) Validation of kinase phosphorylation sites Plk1, Aurora A 32P-ATP and phosphorimaging 358 Identified spindle phosphorylation sites and VTELNDSRSECI and aa scan SPOT (39) Phosphorylation site determination PKA 32P-ATP and phosphorimaging 20mer peptides encompassing intracellular loop of rat cardiac Na+-Ca2+ exchanger NCX1, and substitution analysis of identified substrates SPOT (40) DIPP (41) Kinases: enzymatic activities in cell lysates and kinome profiling Comprehensive PKA, lysate of 192 Kinase peptide 33P-ATP and description of LPS and substrates phosphorimaging phosphorylation events unstimulated induced by LPS PBMCs, stimulation of PBMCs MAPK and inhibitors PD98059, SB203580 Analysis of kinase activities in cell lysate Lysates from M. musculus MAFs and M. musculus MLCECs ± BMK1 knockout 33P-ATP and phosphorimaging 1152 Kinase substrate peptides of mixed species DIPP (42) Fingerprinting effects of DEX on T- Lysate of anti-CD3 and anti-CD28 ab stimulated CD4+ 33P-ATP and phosphorimaging 1176 Peptides containing kinase substrate consensus DIPP (43) lymphocyte kinome cells treated ± DEX sequences Comparison of kinome profiles of Barrett’s esophagus, normal squamous esophagus and normal gastric cardia Tissue lysates 33P-ATP and phosphorimaging 1176 Kinase specific consensus sequences DIPP (44) Investigation of the effect of DEX on adipocyte and Tlymphocyte kinome Lysate of insulin stimulated 3T3 adipocytes treated ± DEX and lysate of anti-CD3 and anti-CD28 ab stimulated CD4+ cells treated ± DEX 33P-ATP and phosphorimaging 1176 Peptides containing kinase substrate consensus sequences DIPP (45) Detection of kinase activity in cell lysate PKA, PKC, MCF-7 lysate incubated ± Forskolin and H89 Biotinylated Phostag followed by DyLight 647 or Cy3 labelled antistreptavidin ab LRRAXLW, YIXGSFK, LRVQNXLRRRR, X = S, pS, T, pT, Y, F, A DIPP (46) Analysis of kinase activities in lysates Lysates from 2 and 5 weeks old A. thaliana and from P. syringae infected A. thaliana 33P-ATP and phosphorimaging 192 Kinase peptide substrates DIPP (47) Screening of renal kinase activities to identify kinases involved in angiotensin II dependent hypertensive renal damage Cortical tissue lysates from sprague dawley rats and Ren 2 rats treated ± ACEi 33P-ATP and phosphorimaging > 1100 Mammalian kinase substrate sequences DIPP (48) Tyrosin kinase profiling in zebrafish embryos Zebrafish embryo lysate (1 day post fertilisation) and knockdown of Src family kinases, Fyn, Yes and Wnt11, Fyn and Yes kinase Fluorescein labelled anti-pTyr ab (PY20, Exalpha Biologicals) 144 Tyr kinase substrate peptides DIPP (49) Tyrosine kinase profiling in zebrafish embryos Zebrafish embryo lysate 3 and 5 days post fertilisation Fluorescein labelled anti-pTyr ab (PY20, Exalpha Biologicals) 144 Tyr kinase substrate peptides DIPP (50) Determination of kinase activities in cell lysates to analyse phylogenetic relations of microarray phosphorylation patterns Lysates from P. pastoris, T. aestivum, C. albicans, A. thaliana, F. solani, D. melanogaster, H. sapiens, M. musculus, D. discoideum and 33P-ATP and phosphorimaging 1152 Kinase substrate peptides of mixed species DIPP (51) Effects of Pichinde virus infection on signalling pathways in C. porcellus macrophages Cytoplasmic macrophages extract after mild and lethal virus infection (P2, P18) 1d and 6d post infection 32P-ATP and phosphorimaging 1176 Peptides containing human in vivo phosphorylation sites DIPP (52) Phosphorylation profiling of A. thaliana proteins Cytosolic and nuclear extracts, cytosolic extracts after stress, SRPK4 (A. thaliana), MPK3 (A. thaliana) 32P-ATP and phosphorimaging 47 Peptides contaning known plant phosphorylation sites DIPP (53) Detection of kinase activities in cell lysates using SPR p60c-Src, PKA, EGFR, MAPKAPK-2, CaMK2, Akt, p38, JNK1, PKC, Erk2, PKC, Abl, InsR, Jak1, PKC and inhibitors SU6656, H-89 and A431 cell lysate ± NGF stimulation and SU6656 addition pTyr detection: anti-pTyr ab (Pt66, Sigma Aldrich) followed by SPR 26 Peptides containing kinase consensus motifs DIPP (54) Kinase profiling of virion components SARS coronavirus lysate 33P-ATP and phosphorimaging 1152 Kinase substrate peptides of mixed species DIPP (55) Fingerprinting of tyrosine kinase activities Lysates of NUP214-Abl and BCR-Abl expressing Ba/F3 cells incubated ± imatinib FITC labelled antipTyr ab (PY20, Exaplha Biologicals) 144 Tyr kinase substrate peptides DIPP (56) Fingerprinting of tyrosine kinase Lysates of human dermal FITC labelled anti- 144 Tyr kinase substrate DIPP (57) pSer/pThr detection: Phostag-biotin followed by streptavidin, anti-streptavidin ab and SPR activities microvascular endothelial cells and of hemangioma endothelia cells pTyr ab (PY20) peptides Analysis of the effect of aspirin and celecoxib on cancer kinome Lysates from colon cancer cell lines DLD1 and HT29 treated with aspirin or with celecoxib 33P-ATP and phosphorimaging 1176 Kinase substrate peptides DIPP (58) Fingerprinting of tyrosine kinase activities Lysates of androgensensitive prostate carcinoma cell line LNCaP FITC labelled antipTyr ab (PY20, Exaplha Biologicals) 144 Tyr kinase substrate peptides DIPP (59) Analysis of the postsynaptic phosphoproteome PKA, CaMK2, Erk2, Rock2, Gsk3, Fes, JNK3, Akt1, PKC, PKC, PKC, p70S6, Rsk2, Erk1, Cdk5, p38, CK1, CK2, c-Raf, MEK1, PDK1, TBK1, Fyn, Pyk2 32P-ATP and phosphorimaging 600 Peptides corresponding to 300 M. musculus in vivo phosphorylation sites in 92 synaptic proteins together with A, V or F variants of the S, T and Y residues and additional peptides containing kinase consensus motifs and a priming array containing 200 peptides from multiple phosphorylated sequences togehter with A, V or F variants of the S, T and Y residues and pS, pT and pY substitutions DIPP (60) Kinome profiling for putative target detection and analysis of differences in primary and metastatic UM lysates Chondrosarcoma lysates from 9 primary cell lines and OUMS27, CH2879, SW1353 and C3842 cell lines and lysates from 2 MSC cultures and 5 colorectal carcinoma cell lines and inhibitors PP1, PP2 and PP3 and Mel270 cell lysate treated 33P-ATP and phosphorimaging 1024 Kinase substrate peptides DIPP (61) ± p60 c-Src siRNA Investigation of the role of phosphorylation in plant sugar response Lysates from A. thaliana treated with water, sorbitol, glucose or sucrose 32P-ATP and phosphorimaging 1176 and 1024 Kinase substrate peptides DIPP (62) Tyrosine kinome profiling Uveal melanoma (UM) cell lysates: 8 primary UM, 3 UM metastases, 3 fresh UM samples and 3 liver metastases FITC labelled antipTyr (PY20, Exalpha Biologicals) 144 Tyr kinase substrate peptides DIPP (63) Kinome profiling Lysates from A. thaliana fed sucrose or sorbitol 32P-ATP and phosphorimaging 960 Kinase consensus sequences selected for their importance in mammalian signal transduction and 1152 kinase substrate peptides of mixed species DIPP (64) Detection of kinase activities in cell lysates p60 c-Src, cytoslic and membrane fraction of MCF-7 lysates, inhibitors PP2 and SU6656,, NGF, CHO and A431 lysates and lysates from M. musculus brain and skin Cy5 anti-pTyr ab 840 Peptides DIPP (65) Kinome profiling for the detection of aberrant kinase activities in neoplastic tissues 29 pediatric brain tissue lysates and lysates from Wilms’ tumors, colon carcinomas, normal kidney and normal colon Fluorescein labelled anti-pTyr ab (PY20, Exalpha Biologicals) 144 Tyr kinase substrate peptides DIPP (66) Kinome analysis of monocytes Lysates of monocytes from B. taurus treated ± LPS 32P-ATP and phosphorimaging 298 Peptides containing proposed B. taurus phosphorylation sites togehter with 2 negative and 7 positive controls DIPP (67) Fingerprinting tyrosine kinase avtivities Lysates of leukemia samples FITC labelled antipTyr ab 144 Tyr kinase substrate peptides DIPP (68) Kinome profiling and B-cell lysates from 88 Systemic 33P-ATP and 1024 Kinase substrate DIPP (69) biomarker identification Lupus Erythematosus patents and from 72 healthy controls phosphorimaging peptides Kinome profiling of osteoblast adhesion MC3T3-E1 cell lysate 2h after adhesion substrate subjection and control 33P-ATP and phosphorimaging 1024 Kinase substrate peptides DIPP (70) Kinome profiling of myxoid liposarcoma Lysates of two myxoid liposarcoma cell lines and primary 33P-ATP and phosphorimaging 1024 Kinase substrate peptides DIPP (71) cultures of four myxoid liposarcomas Screening of kinase activities in response to Jasmonate and salicylate treatment Lysates of A. thaliana incubated with jasmonic acid and/or salicylic acid 33P-ATP and phosphorimaging 1178 Kinase substrate peptides of mixed species DIPP (72) Tumor kinase profiling to predict chemotherapy response Biopsy lysates from rectal cancer patients ± sunitinib Fluorescentlylabelled Anti-pTyr ab 144 Tyr kinase substrate peptides DIPP (73) Fingerprinting tyrosine kinase avtivities Lysates from 4 different cell lines and their matching phosphatase and tensin homolog (PTEN) knockdown sublines Fluorescentlylabelled Anti-pTyr ab 144 Tyr kinase substrate peptides DIPP (74) Fingerprinting tyrosine kinase avtivities Lysates from paediatric brain tumor tissue Fluorescentlylabelled Anti-pTyr ab 144 Tyr kinase substrate peptides DIPP (75) Combinatorial peptide library and interative deconvolution SPOT (76) Kinases: inhibitor identification and characterization Identification of Enzyme I of the 32Ppeptides that inhibit bacterial phosphoenolpyruva enzyme I of the phosphotransferas te and bacterial e system phosphorimaging phosphotransferase system libraries: XXXXXB1HB2XXXXX, XXXXB1HB2XXXX, XXXXXB1CB2XXXXX B = defined, X = randomized Identification of peptidic inhibitors c-GMP dependent kinase 32P- autophosphorylated PKG and phosphorimaging Combinatorial peptide library and interative deconvolution SPOT (77) libraries: XXX12XXX, XXXRK12X, XRKKK12X, 1RKKKKKH12, 12LRKKKKKH 1,2 = defined aa, X = mixture of all aa except Ser, Thr and Ala screening of selected peptides Determination of Ki values for inhibitors p60c-Src and inhibitors quercetin, tyrphostin, A47 and PP1 33P-ATP and surface plasmon resonance (antipTyr ab, BD biosciences) YGEFKKK, FGEFKKK, pYGEFKKK, YAAPKKK, LRRASLG DIPP (78) Investigation of signalling pathways influenced by spongistatin 1 HUVEC lysate treated ± spongistatin 1, combretastatin A4 phosphate or vinblastine 33P-ATP and phosphorimaging 1152 Kinase substrate peptides of mixed species DIPP (79) Prediction of kinase inhibitor resistance in tumors – profiling of repsonse to MTKI of 27 cell lysates to identify signature peptides HT29 lysate transfected with p60 c-Src construct or empty vector incubated ± PP2 or JNJ-26483327 (=MTKI), HCT116 and SKOV-3 lysate with p60 cSrc siRNA, KatoII,SNU-5, MKN45, NSCLC.H441, NCI-N87, SNU484, NSCLCH322 and Caco2 lysates incubated ± JNJ-38877605, FITC labelled antipTyr ab (PY20, Exaplha Biologicals) 144 Tyr kinase substrate peptides DIPP (80) EGF stimulated lysates and lysates from 8 xenograft tumors incubated ± MTKI Analysis of a putative kinase inhibitor Mesenchymal stem cell lysates incubated ± inhibitor PTK787/ZK22258 4 33P-ATP and phosphorimaging 1152 Kinase substrate peptides of mixed species, 12 control peptides DIPP (81) Analysis of the effect of Pazopanib and Lapatinib on A549 kinome A549 cell lysates treated ± Pazopanib or Lapatinib or both HRP conjugate anti-pTyr ab (R&D Systems) 384 Peptides derived from known phosphorylation sites DPCC (82) Effects of antiCD45RB ab on CD45 activity PBMC lysates after incubation with anti-CD45RB ab 33P-ATP and phosphorimaging 1176 Mammalian in vivo kinase consensus peptides DIPP (83) Hydrogel based microarray for assessment of EGFR activity and sensitivity to inhibitors in cell lysates of drug resistant tumors Lysates from NCIH23, NCI-H1650, PC9, H1650-ER, PC9-GR and K562 lysates and inhibitors erlotinib, gefitinib, CL387,785, BIBW2992, CI-1033, PKI-166 Anti-pTyr ab (4610 Upstate), sec. ab HRP labelled AEEEEYFELVAKKK Diff (84) Identification of signalling pathways influenced by flavopiridol HUVEC lysate treated with TNF ± kinase inhibitor flavopiridol 33P-ATP and phosphorimaging 1152 Kinase substrate peptides of mixed species DIPP (85) 32P-ATP and phosphorimaging Combinatorial peptide library and iterative deconvolution: XXX12XXX SPOT (86) SPOT (87) Kinases: assay development/optimization Method for substrate PKA, PKG specificity determination using peptide libraries X = mixture of all 20 aa, 1,2 = individual aa and peptides with different length and Ala or D-Ala variants as putative inhibitors Kinase assay on PKA, PKC, CK1, 32P-ATP and Kinase substrate peptides: RRASVA, cellulose membranes CK2 phosphorimaging QKRPSQRAKYL, DDDDEESITRR, DDDSDDDAAA and Ser to Ala variants, PKA substrates with altering length: AAAARASVA, AARRASVA, RRASVA, RRASVAAA, RRASVAAAAA, RRASCAAAAAA and 9 PKA substrate peptides: LRRASVA, LRRASLG, RRASVA, LRKASLG, LHRASLG, LRHASLG, LKRASLG, LARASLG, LRAASLG Method for substrate specificity determination using peptide libraries PKA 32P-ATP and phosphorimaging Combinatorial peptide library and interative deconvolution SPOT (88) libraries: XXX12XXX, X = mixture of all 20 aa, 1,2 = individual aa Demonstration of applications for protein and peptide microarrays p42 MAPK, PKA, CK2 33P-ATP and phosphorimaging Kemptide, Elk1, protein kinase inhibitor 2 DIPP (89) Demonstration of versatility of peptide arrays for functional assays p60c-Src 33P-ATP and phosphorimaging EEIYGEFF DIPP (90) Development of an antibody based method to detect kinase activity p60c-Src, PKA FITC-labelled phospho-specific abs YIYGSFK, ALRRASLG, YIYGpSFK, ALRRApSLG DIPP (91) Method for detection of weakly interacting protein-protein motifs using paired peptides Erk2 Anti-pElk1 (S383, NEB) ab, HRP labelled sec. Ab KGRKPRDLELP, FWSTLSPIAPR, MNGGAANGRIL SPOT (92) Validation of phosphospecific dye on solid supports Abl ATP or -thio-ATP. Phosphorylation was detected with Pro-Q Diamond dye or thiophosphorylation was detected with thiolreactive Kemptide, Abl tyrosine kinase peptide and Ca2+/calmodulin kinase substrate (glycogen synthase 1-10) in 15 different dilutions on polyacrylamide coated CLPP (93) BODIPY 630/650 methyl bromide reagent slides Method for fluorescence detection of phosphorylation on peptide microarrays Abl, PKA ProQ Diamond (Invitrogen) Abl peptide, CaMK2 peptide, Kemptide DIPP (94) Different methods on peptide arrays for kinase research PKA, CK2, Gsk3, PDK1, Tie2 32P-ATP and phosphorimaging 3 Arrays containing either 710 human phosphorylation sites or 2234 peptides ± priming phosphorylations or 1394 kinase activation loop derived peptides and peptides covering the cytoplasmic domain of Tie2 as overlapping peptide scan DIPP (95) Method to evaluate kinase activities on peptide microarrays using MALDI-TOF/MS p60c-Src, PKA, PKG, CaMK2, CKI, Abl, Erk MALDI-TOF/MS IYGEFKKKC, LRRASLGC, RKRSRAEC, KRQQSFDLFGC, CKRRALpSVASLPGL, IYAAPKKKL, TGPLSPGPFGC DIPP (96) inhibitors Assay to detect phosphorylation using gold nanoparticle probes PKA and inhibitor H89 Biotin-ATP followed by avidin stabilized gold particles, silver enhancement and resonance light scattering LRRASLG, LRRAGLG DIPP (97) Development of a microarray based multi analyte assay which allows incubation with different enzymes in one experiment Abl, p60 c-Src, CK1, Erk1, PKA, PKC, PKG and inhibitors Gleevec, Gö6850 and lysate from K562 cells that overexpress BcrAbl ± Gleevec, peroxide vanadate and calynuclin A treatment MALDI-TOF/MS AIYAAPFKKGC, EEIYGEFE, AKKKC, CKRRALpSVASLPGL, TGPLSPGPFGC, LRRASLGC, AKIQASFRGHMARKKG C, RKRSRAEC, AIpYAAPFKKGC, LRRApSLGC DIPP (98) Method for detection of phosphorylation on peptide microarrays PKA 32P-ATP or biotinylated Zn2+ chelate compound followed by streptavidin and SPR, anti-pSer CGGLRRASLG, Ala substitution and Ser phosphorylated, CGIYGEFKKK and Tyr phosphorylated DIPP (99) (PSR-45, Sigma Aldrich) or anti-pTyr (PT-66, Sigma Aldrich) ab followed by SPR Kinase inhibition assay using sol-gel derived multicomponent microarray PKA and inhibitors H7, H89 ProQ Diamond (Invitrogen) Kemptide Diff (100) Method for screening of kinase – chemical interaction using aerosol deposition of nanodroplets containing the biologically active compound p60c-Src, B-Raf (V599E), KDR, Met, Flt3 (D835Y), Lyn, EGFR, PDGFR, Tie2 and inhibitors staurosporine, PP2, GW5074, KDRI and PDGRF 1-1 Anti-pTyr ab (pTyr100, Cell signalling), sec. Alexa 555 ab Poly(E4-Y)10 peptide DIPP & SPNS (101) Microfluidic peptide microarray applications PKA ProQ Diamond (Invitrogen) R(R/K)XSLG, X = any aa and pY, A and F variants of S LDPS (102) Method for employing kinase inhibition based on labelling phopshorylation events using nanogold particles PKA and Biotin-ATP, detection using avidin stabilized gold nanoparticles followed by silver enhancement and resonance light scattering LRRASLG, LRRAGLG DIPP (103) Solution phase kinase assay CK1, Plk3, Akt1, SGK1, PKC, PKCz, PKA, Aurora A, Aurora B, Chk2, CaMK2, MK2, Chk1, Cdk2, Cdk5, p38, CK2, Abl, Lck, p60c-Src, EphB2, VEGFR2, Kit, Flt3, Jak2, EGFR and inhibitors PKI, SrcI, TBB, staurosporine Fluoresceinylglycin e amide followed by anti-fluorescein ab (Roche), biotinylated sec. ab and strepatavidinAlexa555 or antipTyr-biotin ab (Invitrogen) followed by streptavidin phycoerythrin 900 Peptide substrates encoded by an oligonucleotide sequence composed of annotated kinase substrate sequences and random sequences DCSD (104) Fluorescence based assay for phosphorylation detection PKA, Akt1 and inhibitor staurosporine -elimination followed by Michael addition of thiol containing RPRAASF, LRRASLG SPOT (105) PKA inhibitors H89, HA1077, KN62, mallotoxin tetramethylrhodami ne Protocol for phosphorylation site determination Different human kinases 33P-ATP and phosphorimaging or ProQ Diamond followed by fluorescence reading 3280 Peptides covering all human peptidylprolyl-cis/transisomerases and 17181 peptides covering the CMV proteome DIPP (106) Protocol for kinome profiling using plant tissues, biopsies, cells and purified kinases Plant tissues lysates, biopsie lysates, cells lysate and purified kinases 33P-ATP and phosphorimaging 1176 Kinase substrate peptides from different organisms and a second arrays contaning 1024 in vivo phosphorylation sites DIPP (107) Kinetic studies of PKA catalyzed phosphorylation in real time on a peptide array PKA and inhibitors staurosporine, AMP-PNP and PKA inhibitor peptide Anti-phospho ab, sec. fluorescently labelled ab allowing real time detection of fluorescence using a CCD camera 140 Peptides derived from human phosphorylation sites together with 4 phosphopeptides and detailed phosphorylation analysis of the peptide EILSRRPSYRKIL and kinetic analysis of CREB peptide and Kemptide (LRRASLG) DIPP (108) Assay to identify bisubstrate inhibitors PKC, Anti-pSer, anti-pThr ab (Cell signalling #2351, 9611, 9391), sec. ab FITC labelled 144 Ser/Thr Kinase substrate peptides DIPP (109) Kinase assay using Raman spectroscopy and gold nanoparticle probes PKA and inhibitors H89, HA1077, KN62 Biotinylated antipSer ab (Sigma Aldrich) followed by gold nanoparticles and silver enhancement CGGALRRASLG, CGGALRRAGLG DIPP (110) Assay for the development of a depigmenting assay PKC and inhibitors TMP and bisindoylmaleimid e Anti-pSer/pThr ab, sec. ab Cy5 labelled 30- and 10-meric Peptides as MBP fusion protein derived from tyrosinase Diff (111) Protocol for measurement of kinase activities in cell lysates A431 lysates ± SU6656 incubation, HepG2 lysates and lysates from M. musculus tumors after A431 cell Phos-tag-biotin followed by Dylight 649 conjugated streptavidin or Cy5 labelled anti-pTyr ab (Amersham) Array containing 804 peptides and second array containing peptides EEIpYGEFD, EEIYGEFD, EEIFGEFD DIPP (112) inoculation Protocol for determination of kinase activities in cell lysates HEK293 lysate ProQ Diamond (Invitrogen) 4198 Peptides representing overlapping peptide scans covering all human peptidylprolyl-cis/transisomerases Kinases: peptide immobilization and array fabrication optimization Development of p60c-Src, PKA FITC-labelled YIYGSFK, ALRRASLG, approaches for site phospho-specific KGTGYIKTG, specific immobilisation abs YIYGpSFK, of peptides ALRRApSLG, KGTGpYIKTG DIPP (113) DIPP (114) Review: peptide array applications c-Abl 32P-ATP or fluorescencelabelled anti-pTyr ab (PY20) 720 peptides from annotated human phosphosites DIPP (115) Immobilization strategies for peptides on peptide microarrays PKA, p60c-Src FITC-anti-pTyr and FITC anti-pSer ab (Sigma Aldrich) YIYGSFK, ALRRASLG DIPP (116) Development of an oriented peptide library approach PKA, Cdc15 (S. cerevisiae), ab MPM2, 3F3/2 and PY20 ab 33P-ATP, antiHRP sec. Ab Binding libraries: AXXXXpS/pTXXXXA, AXXXXpYXXXXA, AXXXXXpS/pTQDPFXX A, X ≠ Cys SPOT (117) Kinase library: AXXXXXS/TXXXXA, X ≠ Ser, Thr, Cys Strategy for making peptide arrays using protein-peptide fusions Abl, PKA Cy3/Cy5 labelled anti-pSer and pTyr ab (Sigma Aldrich) Leptin-Kemptide, malic enzyme-Kemptide, Leptin-Abl peptide, malic enzyme-Abl peptide DIPP (118) Immobilization using poly(dT)-modified peptides p60c-Src,PKA and inhibitor AG213 Anti-p-GFAP ab (YC10 Medical and Biochemical Laboratories) and anti-pTyr ab (PY20, Exalpha Biologicals) IYGEFKKK, IAGEFKKK, RRRVTSAARRS, RRRVTAAARRS DIPP (119) Assay for phosphorylation site detection using gold nanoparticle based electrochemical p60 c-Src, PKA and inhibitors ellagic acid, PP2, PP3, genistein, herbimycin A Biotin-ATP followed by streptavidin coated gold nanoparticles and electrochemical Raytide, Kemptide DIPP (120) detection detection Optimized surface chemistry for peptide immobilisation for phosphorylation analysis using different linker moieties p60c-Src, PKA 33P-ATP or SPR imaging using Zn2+ chelate compound CGIYGEFKKK, CGGLRRASLG and modifications DIPP (121) Microarray assembly by accoustic dispensing Pim1 and inhibitor hb1277 32P-ATP and phosphorimaging KKRNRTLTK SPNS (122) Regeneration of phosphorylated SPOT membranes Potato acid phosphatase 32P-ATP and phosphorimaging Kinase substrate peptides: RRASVA, QKRPSQRAKYL, DDDDEESITRR, DDDSDDDAAA and S to A variants, PKA substrates with altering length: AAAARASVA, AARRASVA, RRASVA, RRASVAAA, RRASVAAAAA, RRASCAAAAAA and 9 PKA substrate peptides: LRRASVA, LRRASLG, RRASVA, LRKASLG, LHRASLG, LRHASLG, LKRASLG, LARASLG, LRAASLG SPOT (87) PTP substrate specificity determination PTP1B, PTP, Anti-pTyr ab mixture (4G10, upstate, PY20 and PY69, Transduction Laboratories, pTyr100, Biolabs), HRP sec. Ab AABX1pYX2BAAA, B = mixture of all 20 aa, X1, X2 = individual aa, Tie2 peptides VFSKYpYTPVLA, VFSKYpYpTPVLA, STAT5 peptide MAIESLNYSVYTTNS and pY, pS, pT variants and IR autophosphorylation sequence with all combinations of pY SPOT (123) Substrate identification GST-fusion of substrate-trapping mutants of PTPH1, TCPTP, SAP1, and PTP-1B Radioisotopic labelling of GSTfusion proteins by PKA mediated phosphorylation followed by 7 Peptides derived from cytoplasmic region of GHR in nonphosphorylated and tyrosine phosphorylated form SPOT (124) Phosphatases phosphorimaging Method for detecting weakly interacting protein-protein motifs using paired peptides PTP1B Anti-pTyr ab (Upstate), HRP sec. Ab IR autophosphorylation site IYETDYpYRKGG and overlapping peptides covering the cytoplasmic sequence of IR SPOT (92) Protocol for SPOT applications for PTPs PTP1B Anti-pTyr ab mixture (4G10, upstate, PY20 and PY69, Transduction Laboratories, pTyr100, Biolabs), HRP sec. Ab IR peptide MTRDIYETDYYRKGG and pY substitutions and SPOT double synthesis covering the cytoplasmic IR domain as overlapping peptide scan togehter with the IR autophosphorylation peptide IYETDpYRKGG SPOT (125) Phosphatase profiling using cell lysates K562 cell lysates treated ± Gleevec, peroxide vanadate, calynulin A MALDI-TOF/MS AIYAAPFKKGC, EEIYGEFEAKKKC, CKRRALpSVASLPGL, AIpYAAPFKKGC, LRRApSLGC DIPP (98) Construction of pTyr microarrays for phosphatase specificity determination PTP1B, PTPµ Fluorescence imaging subsequent to treatment with Cy5 labelled anti-pTyr ab 48 pTyr-containing peptides DIPP (126) Assay for substrate specificity determination Lambda Phosphatase, AP, PP1, PP2B, PP2A ± Pin1 ProQ Diamond (Invitrogen) 87 phosphopeptides derived from know phosphorylation sites DIPP (127) Peptide immobilization by click sulfonamide reaction PTP1B Anti-biotin-strepCy5 PDVLEYpYKNEHAK, RDINSLpYEVSRMY DIPP (128) Effects of antiCD45RB ab on CD45 activity PBMC lysates after incubation with anti-CD45RB ab 33P-ATP and phosphorimaging 1176 Mammalian in vivo kinase consensus peptides DIPP (83) Identification of physiological subtrates GST-fusion of substrate-trapping mutant of human Density-Enhanced Phosphatase (CD148) Fluorescence imaging subsequent to incubation with Cy5 labelled anti-GST ab 6057 pTyr-containing peptides derived from human phosphorylation sites and 343 controls DIPP (129) Substrate specificity determination, identification of specific substrates by dualcolor readout Cy3 or Cy5 labelled substratetrapping mutants of PTP-1B, TCPTP, SHP-1, SHP-2, and LMWPTP Fluorescence imaging 144 Putative Tyr phosphatase peptide substrates DIPP (130) Substrate specificity determination PTP1B, TCPTP, SHP1, SHP2, SHP1 and SHP2 catalytic domain, LMWPTP ProQ Diamond (Invitrogen) 144 Putative Tyr phosphatase peptide substrates DIPP (131) Identification of physiological subtrates GST-fusion of substrate-trapping mutant of human PTP-1B Fluorescence imaging subsequent to incubation with Cy5 labelled anti-GST ab 6057 pTyr-containing peptides derived from human phosphorylation sites and 343 controls DIPP (132) Lysin Methyl-Transferases (PKMTs) Determination of the contribution of aa for substrate recognition G9a 3H-AdoMet and tritiumimaging Substitutional analysis of a peptide covering the first 21 aa of H3 (all 20 aa and modified aa) and 45 peptides from putative new substrate proteins SPOT (133) Substrate specificity determination Dim-5 (N. crassa) 3H-AdoMet and tritiumimaging Overlapping peptide scan of the first 21 aa of H3 and K9A variant and substitutional analysis SPOT (134) Substrate specificity determination and identification of novel in vivo substrates Set7/9 3H-AdoMet - 420 peptides representing substitutional analysis of histone H3 tail 1-21 SPOT (135) and tritiumimaging & DPCC - 133 predicted peptides derived from 118 different proteins - 216 peptides derived from histone H3 featuring 18 different posttranslational modifications Peptidyl-Prolyl-cis/trans-Isomerases Identification of Human Pin1 Sequential electroblotting to PVDF- Peptide scan of human Cdc25C including SPOT (136) substrates membranes, incubation of PVDV-membranes with anti-Pin1 ab followed by secondary ab phosphorylated S/T-P moieties Determination of substrate specificity Trigger factor (TF) (E. coli) and TFfragments Sequential electroblotting to PVDFmembranes, incubation of PVDV-membranes with anti-TF ab followed by fluorescence labelled secondary ab Peptide scans (13meric peptides overlapping by 10 residues) of E. coli proteins: elongation factor Tu, methionine biosynthesis enzyme, isocitrate dehydrogenase, glutamine-tRNAsynthetase, alkaline phosphatase, galactosidase, galactose binding protein), L2 (a ribosomal protein), lambda cI, pro-outer membrane protein A, sigma 32, SecA, dehydrofolate reductase and yeast cytochrome B2, F1- and Su9ATPase subunits, Photinus pyralis luciferase, RNaseT1 from Aspergillus oryzae and human PrP (prion protein) SPOT (137) Determination of substrate specificity Trigger factor and DnaK (E. coli) Sequential electroblotting to PVDFmembranes, incubation of PVDV-membranes with anti-TF ab followed by fluorescence labelled secondary ab Peptide scans (13meric peptides overlapping by 10 residues) of E. coli proteins: elongation factor Tu, methionine biosynthesis enzyme, isocitrate dehydrogenase, glutamine-tRNAsynthetase SPOT (138) Identification of Pin1 inhibitors Catalytic domain of human Pin1 Sequential electroblotting to PVDFmembranes, incubation of PVDV-membranes with anti-Pin1 ab followed by secondary ab 1000 pentapeptides of general structure AcXaa-pThr-Yaa-ZaaGln(linker) with Xaa and Zaa = Phe, Abz, Bip, Bth, Cha, Dbg, Nal, tBuGly, tBuPhe, Thi SPOT (139) Yaa = MeAla, MePhe, Pip, Pia, 4Pip, Thz, Tic, Pro, Acc, Amb (all possible combinations) Identification of substrates Human Pin1 ProQ Diamond subsequent to phosphatase treatment 87 phosphopeptides derived from know phosphorylation sites DIPP (127) Arginin Methyl-transferases (RMTs) Substrate screening RMT1, CARM1 3H-AdoMet and tritiumimaging 36 PABP1 and H3 derived peptides containing central Arg SPOT (140) Solution phase assay with tagged substrates allowing selective capturing Protein arginine methyl-transferase 1 (RMT1) MALDI-TOF/MS GGRGGFGC DIPP (141) Consensus motif determination Sumo1 and Hela cell lysate Anti-Sumo1 ab followed by IgG biotin and antistreptavidin-HRP ab 10 RanGAP1 derived peptides containing Sumo consensus motifs and control peptides containing R instead of K for assay optimization, peptides covering 11 Sumo targets and negative control and array containing 640 peptide library for consensus motif determination DIPP & Diff (142) Determination of SUMOylation sites SUMO kit (Biomol) Anti-Sumo1 ab Overlapping peptide scan of PDE4B2, C2, A4, D5 and alanine scan of putative PDE4D5 sites SPOT (143) Ubiquitination kit (Biomol) Anti-ubiquitin ab Overlapping PDE4D5 peptide scan, 6 arrestin 2 derived peptides SPOT (144) ppGalNACT isoform T2 MALDI-TOF/MS GAGAPGPTPGPAGAG K, GAGAPGPSPGPAGAG K, GAPGPTPGPAGK, AcPTPGPAGK, DIPP (145) SUMO-Transferases Ubiquitin-Transferases Determination of ubiquitinylation target sites Glycosyl-transferases Assay for monitoring O-glycosylation GAPGSTAPPAGK, GAHGVTSAPAGK, GAAPDTRPAAGK, YSPTSPSKR, PTTDSTTPAPTTK, DDSIEGSGGR O-glycopeptide library production for disease associated autoantibody screening and substrate specificity comparison GalNAc-T1 and T2 MALDI-TOF/MS, lectin reactivity and mabs 5E5 and 2D9 VTSAPDTRPAPGSTAP PAHG ± GalNAc, PTTTPITTTTTTVTPTPT GTQTPTTTPISTTC, CPPTPSATTPAPPSSS APPETTAA, LSESTTQLPGGGPGCA , MEELGMAPALQPTQG AMPAF, PRFQDSSSSKAPPPSL PSPSPLPG, WKFEHCNFNDYTTRLR ENEL DIPP (146) Glycosylation of printed peptides for peptide diversification for mapping of glycopeptide specific abs GalNAc-T2 and T4 MALDI-TOF/MS Peptides derived from M. musculus podoplanin DIPP (147) Histone Deacetylases (HDACs), Sirtuins (Sirts), ADP-Ribosyl-transferases (ARTs) Detection of ADPribosylation sites pertussis toxin (B. pertussis) 32P NAD+ and phosphorimaging 5- to 25-mer C-terminal peptides of the GProtein -subunits of Gi3, Gi, Gs, Go1, Go2, GoX1, Trod, Gz, Gq/11 and Gh, partial substitutional analysis of the 16-mer C-terminal peptide of the Gi3 -subunit SPOT (148) Determination of substrate specificity Pea HDAC complexes HD1 and HD2 (P. sativum) Free amino groups generated by HDAC action were chemically acetylated, detection of 14Clabelled acetyl residues by autoradiography Overlapping peptides containing KAc from histones H3 and H4 SPOT (149) Analysis of the influence of local and distal substrate HDAC8 MALDI-TOF/MS (SAMDI assay) Peptides derived from the first 19 aa of H4: truncated peptides, R DIPP (150) substitutions, KAc and methylated variants, deletion and extensions variants and substitutions sequences on activity Assay for HDAC profiling HDAC1-3, HDAC8 and HDAC3 togehter with SMRT MALDI-TOF/MS (SAMDI assay) 361 Member acetylated peptide library: GXKAcZGC, X, Z ≠ Cys DIPP (151) Detection of ADPribosylation sites SIR2 (T. brucei) 32P NAD+ and phosphorimaging Overlapping peptide scan of B. taurus H1.1 DIPP (152) Substrate specificity determination and identification of novel in vivo substrates Human Sirt3 Anti-Sirt3 ab followed by HRPconjugated secondary anti-IgG ab 229 Peptides derived from known acetylation sites and 300 lysinecentered 9meric peptides randomly selected from mitochondrial proteome SPOT (153) Substrate specificity determination and identification of isoform specific substrates HDAC8, Sirt1, HDAC2, HDAC3/SMRT, Hela Jurkat and smooth muscle cell lysate and nuclear Hela lysates treated ± thymidine and nocodazole MALDI-TOF/MS (SAMDI assay) 361 Peptides GRKAcXZC, X, Z ≠ Cys and 54 peptides to investigate the function of the Arg residue Nterminally to KAc DIPP (154) Identification of in vivo substrates Human Sirt3 Anti-Sirt3 ab followed by HRPconjugated secondary anti-IgG ab 240 peptides derived from known mitochondrial acetylation sites SPOT (155) Substrate specificity determination Chymotrypsin, papain Fluorescence of released FITClabelled peptide fragment 400 dipeptides Identification of key residues in protease inhibitors Pancreatic elastase (S. scrofa) and third domain of turkey ovomucoid inhibitor (OMTKY3) Chemoluminescenc e mediated by POD fused to protease Substitutional scan of 3 OMTKY3 derived peptides, substitutional analysis and length scan Proteases SPOT SPOT (156) (157) Substrate specificity determination Outer membrane protease T (OmpT) (E.coli), trypsin Fluorescence increase by clavage of internally queched Dap(Dnp)/Abz peptides Library screen: AGXA, P2RRA, AP1RA, ARP-1A, ARRP-2 Identification of protease inhibitors Cathepsin C, cathepsin L Fluorescence imaging of fluorescein labelled PNA-peptide conjugates Putative Cys protease inhibitors fused to PNA derivatives Identification of proteases responsible for transferrin receptor cleavage Purified proteases A80 and A85 (cathepsin G, neutrophil elastase) ± anti neutrophil elastase ab (AHN10) and inhibitors prefabloc SC, aprotinin, elastatinal, E64, phosphoramidon, pepstatin A, EDTA, DMSO Protease SPOT assay – Fluorescence of released Abzlabelled peptide fragment 19 Transferrin receptor derived peptides and substitutions Assay for monitoring of proteolytic activity Caspase 3, Jurkat cell lysate ± granzyme B and inhibitors Fluorescence imaging of fluorescein labelled PNA-peptide conjugates 7 Peptidic acrylates as protease inhibitors fused to PNA derivatives Substrate cleavage site determination Serine protease DegP (E.coli) Protease SPOT assay – Fluorescence of released Abzlabelled peptide fragment Overlapping peptide scans of PapA Determination of substrate specificity Trypsin, gramzyme B, thrombin Fluorescence increase of deacylated aminocoumarine derivative Ac-Leu-Gly-Pro-LysACC-linker, Ac-Nle-ThrPro-Lys-ACC-linker, and Ac-Ile-Glu-Pro-AspACC-linker or 361member library of general structure Ac-AlaXaa1-Xaa2-Lys-ACClinker with Xaa1=Xaa2= all proteinogenic amino acids except cysteine) SPOT DCSD SPOT DCSD SPOT DIPP (158) (159) (160) (161) (162) (163) Protease assay using microarrays containing glycerol nanodroplets and aerosol deposited substances Thrombin, chymotrypsin and caspase 2 ± corresponding CHO-inhibitors, thrombin ± benzamidine, plasminogen activator, urokinase, factor Xa, plasmin, kallikrein and caspases 2, 4, 6 for compound library incubation ± VEID-MCA or LEHD-MCA Fluorescence of released methoxycoumarylamine Boc-VPR-MCA and 2 BODIPY labelled casein substrates, peptide-MCA substrates: VEID, LEHD, YVAD, DEVD, VDVAD, IETD and 352 compound library Method for solutionbased screening of protease substrate specificity Caspase 3, thrombin, plasmin and apoptotic and non apoptotic cell lysate Fluorescence imaging, rhodamine-labelled protease substrate linked to PNA, NleTRP, DEVD and 192 member split and mix tetrapeptide library: R1R2R3R4, R1 = D, R, L, R2 = F, V, T, P, R3 = D, R, T, P, R4 = D, R, P, Nle, spatial deconvolution by oligopeptide microarray hybridization Protease activity measurement using semi-wet supramolecular hydrogel Lysyl endopeptidase, chymotrypsin, V8 protease and inhibitors Fluorescence increase of released DANSyl derivative SSSSK-linker-DANSyl, SSSSE-linker-DANSyl, DANSyl-FKSSKS Determination of substrate requirements and development of a hK2 avtivated prodrug for prostate cancer therapy Glandular kallikrein 2 (hK2), trypsin Protease SPOT assay – Fluorescence of released Abzlabelled peptide fragment Semenogelin I and II derived peptides with Nterm. Abz, for the best two substrates aa substitutions in P1’ and Lys or His substitutions in P1 and fluorescence quenched combinatorial library with radom sequences in P3-P1: YIGKAXXX, X ≠ Cys Method using real-time SPR imaging to obtain multiplexed kinetic informations and analysis of the sequence specificity of Factor Xa real-time SPR imaging (anti-Flag ab) Prothrombin factor Xa recognition sequences CSGIEGRDYKDDDDK, CSGIEGADYKDDDDK, CSGDYKDDDDK SPNS DCSD Diff SPOT DIPP (164) (165) (166) (167) (168) factor Xa Substrate specificity determination APC, kallikrein, factor VIIa and TF, IXa, XIa, XIIa, complement C1r, C1s, complement factor D, trypsin, tryptase, subtilisin carlsberg, cathepsin B, G, H, K, L, S, V, rhodesain, papain, chymopapain, ficain, stem bromelain Fluorescence increase of released aminocoumarin derivative 722 Member library: P4P3P2P1, P4 = Ala, P3, P2 ≠ Cys, P1 = Lys, Arg Substrate specificity determination Thrombin, plasmin, factor Xa, uPA, thrombin (B. taurus, S. salmothymus) Fluorescence increase of released aminocoumarin derivative 722 Member library: P4P3P2P1, P4 = Ala, P3, P2 ≠ Cys, P1 = Lys, Arg Identification of optimized substrates for blood proteases Kallikrein, factor Xa, thrombin, uPA, plasmin and plasma treated ± Ca2+, tissue factor, kaolin, uPA and 10 plasma samples and 10 proteases (kallikrein, factor Xa, XIIa, XIa, IXa, VIIa, thrombin, APC, uPA, plasmin) Fluorescence of released methoxycoumarylamine Z-FR-MCA, Boc-IEGRMCA, Boc-VPR-MCA, glutaryl-GR-MCA, BocVLK-MCA and 361 compound library: AcXXXX-ACC Identification of substrates ADAM8 Protease SPOT assay – Fluorescence of released Abzlabelled peptide fragment 34 Peptides derived from proteins involved in inflammatory processes and immune response in the nervous system and various peptide derived from MBP cleavage sites Analysis of protease cleavage specificity Chymopapain, subtilisin Fluorescence increase by clavage of internally queched peptides(FAM/TAM RA) 10000 Member PNA encoded library: TAMRA-ßAla-Ser-Xaa4Xaa3-Xaa2-Xaa1-AlaLys(FAM)-TtdsLys(PNA)-amide with TAMRA = Quencher/Normalization SPNS SPNS SPNS SPOT DCSD (169) (170) (171) (172) (173) fluorophor; FAM = Fluorophor, Ttds = PEGbased-linker moiety, Xaa = random positions with amino acid residues Ala, Asp, Phe, Lys, Leu, Asn, Pro, Ser, Val, Tyr. Monitoring caspase activity Purified caspase 3 or lysates of CHO cells treated with staurosporin Fluorescence imaging subsequent to protease treatment ADEVDA and negative control AEVEEA flanked by Cy3 fluorophore and QSY7-quencher moiety. Positive control without quencher Assay for measuring caspase activities in cell lysates Caspase 3 and 8, SKW6.4 and Jurkat (caspase 8 or deficient) lysate treated ± LzCD95L or staurosporine MALDI-TOF/MS (SAMDI assay) DEDVAFC, IETDAFC, CGGDEVDSG, CGDEVDSGVDEVA, CGKRKGDEVDSG, CGGIETDSG, CDGIETDSG, CDGIETDSGVDDD, CGELDSGIETDSG Surface optimization for SPOT synthesis Chymotrypsin fluorescence imaging subsequent to treatment with Cy5labelled streptavidin Biotinylated peptides of general structure biotinGA-P1-G-linker with P1= A,F, G, I, L, P, V, W SPR based assay for monitoring protease activities Caspase 3, 6, 8, CHO lysate incubated ± staurosporine and inhibitor DEVDCHO Surface plasmon resonance imaging subsequent to treatment with protease followed by streptavidin Caspase substrates DEVD, VEID, IETD and negative controls EVEE, VIEE, TIEE, respectively, flanked by cysteinyl-PEG-linker Nterminally and by GGSK(biotinyl)-amide Cterminally Multiplexed solutionphase assay for protease profiling Trypsin, chymotrypsin, endoproteinases Asp-N, Glu-C, Lys-C, Arg-C, thrombin, factor Xa, HRVI, caspases, MMPs, enterokinases, tobacco etch virus protease and Jurkat cell lysate incubated ± Fas Protease cleavage separates His-tag from biotin residue allowing removal of uncleaved peptides by streptavidin beads, readout using anti-His-tag ab Over 1000 DNA encoded, biotinylated peptide derivatives with pentaHis-tag representing substrates for several protease groups DIPP DIPP SPOT DIPP DCSD (174) (175) (176) (177) (178) ab or Bcl2 overexpressing Jurkat lysate Design of biosensors Botulinium neurotoxin and light chain of botulinium neurotoxin FRET activity assay yielding fluorescence increase subsequent to proteolytic cleavage SNAP-25 peptide substrate (FITC/DABCYL labelled) Substrate specificity determination Subtilase 3 S. lycopersicum Protease SPOT assay – Fluorescence of released Abzlabelled peptide fragment 72 Peptides derived from systemin and substitution analysis using A, F, P, S, N, D, K, V, L, and H for P5 to P3´position SPOT (180) Validation of protease assay Caspase 3, caspase 7, chymotrypsin, subtilisin, thrombin, trypsin Fluorescence increase of deacylated rhodamine derivative 10 Peptide sequences derived from known substrates C-terminally linked to fluorophore DIPP (181) Substrate specificity comparison Granzyme A, granzyme K and inactive variants Fluorescence decrease subsequent to treatment with fluorescein labelled streptavidin 1000 Biotinylated random peptides and 24 controls DIPP (182) Monitoring of proteolytic activities onto fluorous based microarrays Thrombin, plasmin, chymotrypsin, trypsin, granzyme B Fluorescence increase of deacylated aminocoumarine derivative Bz-FVR-ACC, Bz-VPRACC, Ac-IEPD-ACC, Cbz-FR-ACC, Suc-AFKACC; Suc-AAPF-ACC, Bz-FVR-AMC, Bz-VPRAMC, AC-IEPD-AMC, Cbz-FR-AMC, Suc-AFKAMC, Suc-AAPF-AMC DIPP (183) Assay for hydrolytic enzymes (proteases, esterases, epoxid hydrolases, phosphatases) Epoxide hydrolase (R. rhodochrous), acetylcholin esterase (E. electricus), trypsin (B. taurus), AP (B. taurus) Enzyme recognition head coupled to fluorogenic moiety which serves as a reporter group that translates enzymatic activity into fluorescence readouts Asp and Lys derivatives DIPP (184) Novel based assay for identification of HRP, AP, -Gal Fluorescently labelled substrate 10000 Random peptide DIPP (185) Diff (179) Miscellaneous modulators of enzyme function analoga sequences CLPP = Cross Linking of Pre-synthesized Peptides DCSD = DNA /PNA Chips as Sorting Device Diff. = Different technologies DIPP = Directed Immobilization of Pre-synthesized Peptides DPCC = Deposition of Peptide-Cellulose Conjugates (Celluspots) LDPS = Light Directed Peptide Synthesis SODA = Synthesis On Defined Areas SPNS = Solution Phase and Nano Spray SPOT = SPOT Synthesis of Peptides References 1. Luo, K., Zhou, P. and Lodish, H.F. (1995) The specificity of the transforming growth factor beta receptor kinases determined by a spatially addressable peptide library. Proc Natl Acad Sci U S A, 92, 11761-11765. 2. oomik, R. and Ek, P. (1997) A potent and highly selective peptide substrate for protein kinase C assay. Biochem J, 322 ( Pt 2), 455-460. 3. Moilanen, A.M., Karvonen, U., Poukka, H., Janne, O.A. and Palvimo, J.J. (1998) Activation of androgen receptor function by a novel nuclear protein kinase. Mol Biol Cell, 9, 2527-2543. 4. Dostmann, W.R., Nickl, C., Thiel, S., Tsigelny, I., Frank, R. and Tegge, W.J. (1999) Delineation of selective cyclic GMP-dependent protein kinase Ialpha substrate and inhibitor peptides based on combinatorial peptide libraries on paper. Pharmacol Ther, 82, 373-387. 5. Himpel, S., Tegge, W., Frank, R., Leder, S., Joost, H.G. and Becker, W. (2000) Specificity determinants of substrate recognition by the protein kinase DYRK1A. J Biol Chem, 275, 24312438. 6. Loog, M., Toomik, R., Sak, K., Muszynska, G., Jarv, J. and Ek, P. (2000) Peptide phosphorylation by calcium-dependent protein kinase from maize seedlings. Eur J Biochem, 267, 337-343. 7. Dostmann, W.R., Tegge, W., Frank, R., Nickl, C.K., Taylor, M.S. and Brayden, J.E. (2002) Exploring the mechanisms of vascular smooth muscle tone with highly specific, membrane-permeable inhibitors of cyclic GMP-dependent protein kinase Ialpha. Pharmacol Ther, 93, 203-215. 8. Lizcano, J.M., Deak, M., Morrice, N., Kieloch, A., Hastie, C.J., Dong, L., Schutkowski, M., Reimer, U. and Alessi, D.R. (2002) Molecular basis for the substrate specificity of NIMA-related kinase-6 (NEK6). Evidence that NEK6 does not phosphorylate the hydrophobic motif of ribosomal S6 protein kinase and serum- and glucocorticoid-induced protein kinase in vivo. J Biol Chem, 277, 27839-27849. 9. Uttamchandani, M., Chan, E.W., Chen, G.Y. and Yao, S.Q. (2003) Combinatorial peptide microarrays for the rapid determination of kinase specificity. Bioorg Med Chem Lett, 13, 29973000. 10. Panse, S., Dong, L., Burian, A., Carus, R., Schutkowski, M., Reimer, U. and Schneider-Mergener, J. (2004) Profiling of generic anti-phosphopeptide antibodies and kinases with peptide microarrays using radioactive and fluorescence-based assays. Mol Divers, 8, 291-299. 11. Rychlewski, L., Kschischo, M., Dong, L., Schutkowski, M. and Reimer, U. (2004) Target specificity analysis of the Abl kinase using peptide microarray data. J Mol Biol, 336, 307-311. 12. Bucko-Justyna, M., Lipinski, L., Burgering, B.M. and Trzeciak, L. (2005) Characterization of testisspecific serine-threonine kinase 3 and its activation by phosphoinositide-dependent kinase-1dependent signalling. FEBS J, 272, 6310-6323. 13. Wang, H. and Brautigan, D.L. (2006) Peptide microarray analysis of substrate specificity of the transmembrane Ser/Thr kinase KPI-2 reveals reactivity with cystic fibrosis transmembrane conductance regulator and phosphorylase. Mol Cell Proteomics, 5, 2124-2130. 14. Papadopoulos, C., Arato, K., Lilienthal, E., Zerweck, J., Schutkowski, M., Chatain, N., MullerNewen, G., Becker, W. and de la Luna, S. (2010) Splice variants of the dual-specificity tyrosine phosphorylation-regulated kinase 4 (DYRK4) differ in their subcellular localization and catalytic activity. J Biol Chem. 15. Beullens, M., Vancauwenbergh, S., Morrice, N., Derua, R., Ceulemans, H., Waelkens, E. and Bollen, M. (2005) Substrate specificity and activity regulation of protein kinase MELK. J Biol Chem, 280, 40003-40011. 16. Mah, A.S., Elia, A.E., Devgan, G., Ptacek, J., Schutkowski, M., Snyder, M., Yaffe, M.B. and Deshaies, R.J. (2005) Substrate specificity analysis of protein kinase complex Dbf2-Mob1 by peptide library and proteome array screening. BMC Biochem, 6, 22. 17. Pouchain, D., Diaz-Mochon, J.J., Bialy, L. and Bradley, M. (2007) A 10,000 member PNA-encoded peptide library for profiling tyrosine kinases. ACS Chem Biol, 2, 810-818. 18. Stulemeijer, I.J., Stratmann, J.W. and Joosten, M.H. (2007) Tomato mitogen-activated protein kinases LeMPK1, LeMPK2, and LeMPK3 are activated during the Cf-4/Avr4-induced hypersensitive response and have distinct phosphorylation specificities. Plant Physiol, 144, 1481-1494. 19. Leung, G.C., Ho, C.S., Blasutig, I.M., Murphy, J.M. and Sicheri, F. (2007) Determination of the Plk4/Sak consensus phosphorylation motif using peptide spots arrays. FEBS Lett, 581, 77-83. 20. Warner, N., Wybenga-Groot, L.E. and Pawson, T. (2008) Analysis of EphA4 receptor tyrosine kinase substrate specificity using peptide-based arrays. FEBS J, 275, 2561-2573. 21 Vilk, G., Weber, J.E., Turowec, J.P., Duncan, J.S., Wu, C., Derksen, D.R., Zien, P., Sarno, S., Donella-Deana, A., Lajoie, G. et al. (2008) Protein kinase CK2 catalyzes tyrosine phosphorylation in mammalian cells. Cell Signal, 20, 1942-1951. 22 Merckx, A., Echalier, A., Langford, K., Sicard, A., Langsley, G., Joore, J., Doerig, C., Noble, M. and Endicott, J. (2008) Structures of P. falciparum protein kinase 7 identify an activation motif and leads for inhibitor design. Structure, 16, 228-238. 23 Parikh, K., Diks, S.H., Tuynman, J.H., Verhaar, A., Lowenberg, M., Hommes, D.W., Joore, J., Pandey, A. and Peppelenbosch, M.P. (2009) Comparison of peptide array substrate phosphorylation of c-Raf and mitogen activated protein kinase kinase kinase 8. PLoS One, 4, e6440. 24 Bohmer, F.D. and Uecker, A. (2009) A substrate peptide for the FLT3 receptor tyrosine kinase. Br J Haematol, 144, 127-130. 25 Han, X., Sonoda, T., Mori, T., Yamanouchi, G., Yamaji, T., Shigaki, S., Niidome, T. and Katayama, Y. (2010) Protein kinase substrate profiling with a high-density peptide microarray. Comb Chem High Throughput Screen, 13, 777-789. 26 Miller, M., Donat, S., Rakette, S., Stehle, T., Kouwen, T.R., Diks, S.H., Dreisbach, A., Reilman, E., Gronau, K., Becher, D. et al. (2010) Staphylococcal PknB as the First Prokaryotic Representative of the Proline-Directed Kinases. PLoS One, 5, e9057. 27 Fraser, J.A., Vojtesek, B. and Hupp, T.R. (2010) A Novel p53 Phosphorylation Site within the MDM2 Ubiquitination Signal: I. PHOSPHORYLATION AT SER269 IN VIVO IS LINKED TO INACTIVATION OF p53 FUNCTION. J Biol Chem, 285, 37762-37772. 28 Galello, F., Portela, P., Moreno, S. and Rossi, S. (2010) Characterization of substrates that have a differential effect on Saccharomyces cerevisiae protein kinase A holoenzyme activation. J Biol Chem, 285, 29770-29779. 29 Szallasi, Z., Denning, M.F., Chang, E.Y., Rivera, J., Yuspa, S.H., Lehel, C., Olah, Z., Anderson, W.B. and Blumberg, P.M. (1995) Development of a rapid approach to identification of tyrosine phosphorylation sites: application to PKC delta phosphorylated upon activation of the high affinity receptor for IgE in rat basophilic leukemia cells. Biochem Biophys Res Commun, 214, 888-894. 30 Edlund, M., Wikstrom, K., Toomik, R., Ek, P. and Obrink, B. (1998) Characterization of protein kinase C-mediated phosphorylation of the short cytoplasmic domain isoform of C-CAM. FEBS Lett, 425, 166-170. 31 Buss, H., Dorrie, A., Schmitz, M.L., Frank, R., Livingstone, M., Resch, K. and Kracht, M. (2004) Phosphorylation of serine 468 by GSK-3beta negatively regulates basal p65 NF-kappaB activity. J Biol Chem, 279, 49571-49574. 32 Carnegie, G.K., Smith, F.D., McConnachie, G., Langeberg, L.K. and Scott, J.D. (2004) AKAP-Lbc nucleates a protein kinase D activation scaffold. Mol Cell, 15, 889-899. 33 Li, Y., Keller, D.M., Scott, J.D. and Lu, H. (2005) CK2 phosphorylates SSRP1 and inhibits its DNA-binding activity. J Biol Chem, 280, 11869-11875. 34 Collins, M.O., Yu, L., Coba, M.P., Husi, H., Campuzano, I., Blackstock, W.P., Choudhary, J.S. and Grant, S.G. (2005) Proteomic analysis of in vivo phosphorylated synaptic proteins. J Biol Chem, 280, 5972-5982. 35 Wiesner, S., Wybenga-Groot, L.E., Warner, N., Lin, H., Pawson, T., Forman-Kay, J.D. and Sicheri, F. (2006) A change in conformational dynamics underlies the activation of Eph receptor tyrosine kinases. EMBO J, 25, 4686-4696. 36 Delgado, J.Y., Coba, M., Anderson, C.N., Thompson, K.R., Gray, E.E., Heusner, C.L., Martin, K.C., Grant, S.G. and O'Dell, T.J. (2007) NMDA receptor activation dephosphorylates AMPA receptor glutamate receptor 1 subunits at threonine 840. J Neurosci, 27, 13210-13221. 37 Amanchy, R., Zhong, J., Molina, H., Chaerkady, R., Iwahori, A., Kalume, D.E., Gronborg, M., Joore, J., Cope, L. and Pandey, A. (2008) Identification of c-Src tyrosine kinase substrates using mass spectrometry and peptide microarrays. J Proteome Res, 7, 3900-3910. 38 Yu, J.C., Chen, J.R., Lin, C.H., Zhang, G., Lam, P.S., Wenger, K.H., Mozaffari, F.B., Huang, S.T. and Borke, J.L. (2009) Tensile strain-induced Ets-2 phosphorylation by CaMKII and the homeostasis of cranial sutures. Plast Reconstr Surg, 123, 83S-93S. 39 Santamaria, A., Wang, B., Elowe, S., Malik, R., Zhang, F., Bauer, M., Schmidt, A., Sillje, H.H., Koerner, R. and Nigg, E.A. The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol Cell Proteomics. 40 Wanichawan, P., Louch, W.E., Hortemo, K.H., Austbo, B., Lunde, P.K., Scott, J.D., Sejersted, O.M. and Carlson, C.R. (2011) Full length cardiac Na+-Ca2+ exchanger (NCX1) protein is not phosphorylated by protein kinase A (PKA). Am J Physiol Cell Physiol. 41 Diks, S.H., Kok, K., O'Toole, T., Hommes, D.W., van Dijken, P., Joore, J. and Peppelenbosch, M.P. (2004) Kinome profiling for studying lipopolysaccharide signal transduction in human peripheral blood mononuclear cells. J Biol Chem, 279, 49206-49213. 42 Hayashi, M., Fearns, C., Eliceiri, B., Yang, Y. and Lee, J.D. (2005) Big mitogen-activated protein kinase 1/extracellular signal-regulated kinase 5 signaling pathway is essential for tumor-associated angiogenesis. Cancer Res, 65, 7699-7706. 43 Lowenberg, M., Tuynman, J., Bilderbeek, J., Gaber, T., Buttgereit, F., van Deventer, S., Peppelenbosch, M. and Hommes, D. (2005) Rapid immunosuppressive effects of glucocorticoids mediated through Lck and Fyn. Blood, 106, 1703-1710. 44 van Baal, J.W., Diks, S.H., Wanders, R.J., Rygiel, A.M., Milano, F., Joore, J., Bergman, J.J., Peppelenbosch, M.P. and Krishnadath, K.K. (2006) Comparison of kinome profiles of Barrett's esophagus with normal squamous esophagus and normal gastric cardia. Cancer Res, 66, 11605-11612. 45 Lowenberg, M., Tuynman, J., Scheffer, M., Verhaar, A., Vermeulen, L., van Deventer, S., Hommes, D. and Peppelenbosch, M. (2006) Kinome analysis reveals nongenomic glucocorticoid receptor-dependent inhibition of insulin signaling. Endocrinology, 147, 35553562. 46 Shigaki, S., Yamaji, T., Han, X., Yamanouchi, G., Sonoda, T., Okitsu, O., Mori, T., Niidome, T. and Katayama, Y. (2007) A peptide microarray for the detection of protein kinase activity in cell lysate. Anal Sci, 23, 271-275. 47 Ritsema, T., Joore, J., van Workum, W. and Pieterse, C.M. (2007) Kinome profiling of Arabidopsis using arrays of kinase consensus substrates. Plant Methods, 3, 3. 48 de Borst, M.H., Diks, S.H., Bolbrinker, J., Schellings, M.W., van Dalen, M.B., Peppelenbosch, M.P., Kreutz, R., Pinto, Y.M., Navis, G. and van Goor, H. (2007) Profiling of the renal kinome: a novel tool to identify protein kinases involved in angiotensin II-dependent hypertensive renal damage. Am J Physiol Renal Physiol, 293, F428-437. 49 Lemeer, S., Jopling, C., Naji, F., Ruijtenbeek, R., Slijper, M., Heck, A.J. and den Hertog, J. (2007) Protein-tyrosine kinase activity profiling in knock down zebrafish embryos. PLoS ONE, 2, e581. 50 Lemeer, S., Ruijtenbeek, R., Pinkse, M.W., Jopling, C., Heck, A.J., den Hertog, J. and Slijper, M. (2007) Endogenous phosphotyrosine signaling in zebrafish embryos. Mol Cell Proteomics, 6, 2088-2099. 51 Diks, S.H., Parikh, K., van der Sijde, M., Joore, J., Ritsema, T. and Peppelenbosch, M.P. (2007) Evidence for a minimal eukaryotic phosphoproteome? PLoS ONE, 2, e777. 52 Bowick, G.C., Fennewald, S.M., Scott, E.P., Zhang, L., Elsom, B.L., Aronson, J.F., Spratt, H.M., Luxon, B.A., Gorenstein, D.G. and Herzog, N.K. (2007) Identification of differentially activated cell-signaling networks associated with pichinde virus pathogenesis by using systems kinomics. J Virol, 81, 1923-1933. 53 de la Fuente van Bentem, S., Anrather, D., Dohnal, I., Roitinger, E., Csaszar, E., Joore, J., Buijnink, J., Carreri, A., Forzani, C., Lorkovic, Z.J. et al. (2008) Site-specific phosphorylation profiling of Arabidopsis proteins by mass spectrometry and peptide chip analysis. J Proteome Res, 7, 2458-2470. 54 Mori, T., Inamori, K., Inoue, Y., Han, X., Yamanouchi, G., Niidome, T. and Katayama, Y. (2008) Evaluation of protein kinase activities of cell lysates using peptide microarrays based on surface plasmon resonance imaging. Anal Biochem, 375, 223-231. 55 Neuman, B.W., Joseph, J.S., Saikatendu, K.S., Serrano, P., Chatterjee, A., Johnson, M.A., Liao, L., Klaus, J.P., Yates, J.R., 3rd, Wuthrich, K. et al. (2008) Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J Virol, 82, 5279-5294. 56 De Keersmaecker, K., Versele, M., Cools, J., Superti-Furga, G. and Hantschel, O. (2008) Intrinsic differences between the catalytic properties of the oncogenic NUP214-ABL1 and BCR-ABL1 fusion protein kinases. Leukemia, 22, 2208-2216. 57 Jinnin, M., Medici, D., Park, L., Limaye, N., Liu, Y., Boscolo, E., Bischoff, J., Vikkula, M., Boye, E. and Olsen, B.R. (2008) Suppressed NFAT-dependent VEGFR1 expression and constitutive VEGFR2 signaling in infantile hemangioma. Nat Med, 14, 1236-1246. 58 Tuynman, J.B., Vermeulen, L., Boon, E.M., Kemper, K., Zwinderman, A.H., Peppelenbosch, M.P. and Richel, D.J. (2008) Cyclooxygenase-2 inhibition inhibits c-Met kinase activity and Wnt activity in colon cancer. Cancer Res, 68, 1213-1220. 59 Bratland, A., Boender, P.J., Hoifodt, H.K., Ostensen, I.H., Ruijtenbeek, R., Wang, M.Y., Berg, J.P., Lilleby, W., Fodstad, O. and Ree, A.H. (2009) Osteoblast-induced EGFR/ERBB2 signaling in androgen-sensitive prostate carcinoma cells characterized by multiplex kinase activity profiling. Clin Exp Metastasis, 26, 485-496. 60 Coba, M.P., Pocklington, A.J., Collins, M.O., Kopanitsa, M.V., Uren, R.T., Swamy, S., Croning, M.D., Choudhary, J.S. and Grant, S.G. (2009) Neurotransmitters drive combinatorial multistate postsynaptic density networks. Sci Signal, 2, ra19. 61 Schrage, Y.M., Briaire-de Bruijn, I.H., de Miranda, N.F., van Oosterwijk, J., Taminiau, A.H., van Wezel, T., Hogendoorn, P.C. and Bovee, J.V. (2009) Kinome profiling of chondrosarcoma reveals SRC-pathway activity and dasatinib as option for treatment. Cancer Res, 69, 62166222. 62 Ritsema, T., Brodmann, D., Diks, S.H., Bos, C.L., Nagaraj, V., Pieterse, C.M., Boller, T., Wiemken, A. and Peppelenbosch, M.P. (2009) Are small GTPases signal hubs in sugarmediated induction of fructan biosynthesis? PLoS One, 4, e6605. 63 Maat, W., el Filali, M., Dirks-Mulder, A., Luyten, G.P., Gruis, N.A., Desjardins, L., Boender, P., Jager, M.J. and van der Velden, P.A. (2009) Episodic Src activation in uveal melanoma revealed by kinase activity profiling. Br J Cancer, 101, 312-319. 64 Ritsema, T. and Peppelenbosch, M.P. (2009) Kinome profiling of sugar signaling in plants using multiple platforms. Plant Signal Behav, 4, 1169-1173. 65 Han, X., Yamanouchi, G., Mori, T., Kang, J.H., Niidome, T. and Katayama, Y. (2009) Monitoring protein kinase activity in cell lysates using a high-density peptide microarray. J Biomol Screen, 14, 256-262. 66 Sikkema, A.H., Diks, S.H., den Dunnen, W.F., ter Elst, A., Scherpen, F.J., Hoving, E.W., Ruijtenbeek, R., Boender, P.J., de Wijn, R., Kamps, W.A. et al. (2009) Kinome profiling in pediatric brain tumors as a new approach for target discovery. Cancer Res, 69, 5987-5995. 67 Jalal, S., Arsenault, R., Potter, A.A., Babiuk, L.A., Griebel, P.J. and Napper, S. (2009) Genome to kinome: species-specific peptide arrays for kinome analysis. Sci Signal, 2, pl1. 68 Ter Elst, A., Diks, S.H., Kampen, K.R., Hoogerbrugge, P.M., Ruijtenbeek, R., Boender, P.J., Sikkema, A.H., Scherpen, F.J., Kamps, W.A., Peppelenbosch, M.P. et al. (2010) Identification of new possible targets for leukemia treatment by kinase activity profiling. Leuk Lymphoma. 69 Taher, T.E., Parikh, K., Flores-Borja, F., Mletzko, S., Isenberg, D.A., Peppelenbosch, M.P. and Mageed, R.A. (2010) Protein phosphorylation and kinome profiling reveal altered regulation of multiple signaling pathways in B lymphocytes from patients with systemic lupus erythematosus. Arthritis Rheum, 62, 2412-2423. 70 Milani, R., Ferreira, C.V., Granjeiro, J.M., Paredes-Gamero, E.J., Silva, R.A., Justo, G.Z., Nader, H.B., Galembeck, E., Peppelenbosch, M.P., Aoyama, H. et al. (2010) Phosphoproteome reveals an atlas of protein signaling networks during osteoblast adhesion. J Cell Biochem. 71 Willems, S.M., Schrage, Y.M., Bruijn, I.H., Szuhai, K., Hogendoorn, P.C. and Bovee, J.V. (2010) Kinome profiling of myxoid liposarcoma reveals NF-kappaB-pathway kinase activity and casein kinase II inhibition as a potential treatment option. Mol Cancer, 9, 257. 72 Ritsema, T., van Zanten, M., Leon-Reyes, A., Voesenek, L.A., Millenaar, F.F., Pieterse, C.M. and Peeters, A.J. (2010) Kinome Profiling Reveals an Interaction Between Jasmonate, Salicylate and Light Control of Hyponastic Petiole Growth in Arabidopsis thaliana. PLoS One, 5, e14255. 73 Folkvord, S., Flatmark, K., Dueland, S., de Wijn, R., Groholt, K.K., Hole, K.H., Nesland, J.M., Ruijtenbeek, R., Boender, P.J., Johansen, M. et al. (2010) Prediction of response to preoperative chemoradiotherapy in rectal cancer by multiplex kinase activity profiling. Int J Radiat Oncol Biol Phys, 78, 555-562. 74 Vivanco, I., Rohle, D., Versele, M., Iwanami, A., Kuga, D., Oldrini, B., Tanaka, K., Dang, J., Kubek, S., Palaskas, N. et al. (2010) The phosphatase and tensin homolog regulates epidermal growth factor receptor (EGFR) inhibitor response by targeting EGFR for degradation. Proc Natl Acad Sci U S A, 107, 6459-6464. 75 Sikkema, A.H., de Bont, E.S., Molema, G., Dimberg, A., Zwiers, P.J., Diks, S.H., Hoving, E.W., Kamps, W.A., Peppelenbosch, M.P. and den Dunnen, W.F. (2011) VEGFR-2 signalling activity in paediatric pilocytic astrocytoma is restricted to tumour endothelial cells. Neuropathol Appl Neurobiol. 76 Mukhija, S., Germeroth, L., Schneider-Mergener, J. and Erni, B. (1998) Identification of peptides inhibiting enzyme I of the bacterial phosphotransferase system using combinatorial cellulose-bound peptide libraries. Eur J Biochem, 254, 433-438. 77 Dostmann, W.R., Taylor, M.S., Nickl, C.K., Brayden, J.E., Frank, R. and Tegge, W.J. (2000) Highly specific, membrane-permeant peptide blockers of cGMP-dependent protein kinase Ialpha inhibit NO-induced cerebral dilation. Proc Natl Acad Sci U S A, 97, 14772-14777. 78 Houseman, B.T., Huh, J.H., Kron, S.J. and Mrksich, M. (2002) Peptide chips for the quantitative evaluation of protein kinase activity. Nat Biotechnol, 20, 270-274. 79 Rothmeier, A.S., Ischenko, I., Joore, J., Garczarczyk, D., Furst, R., Bruns, C.J., Vollmar, A.M. and Zahler, S. (2009) Investigation of the marine compound spongistatin 1 links the inhibition of PKCalpha translocation to nonmitotic effects of tubulin antagonism in angiogenesis. FASEB J, 23, 1127-1137. 80 Versele, M., Talloen, W., Rockx, C., Geerts, T., Janssen, B., Lavrijssen, T., King, P., Gohlmann, H.W., Page, M. and Perera, T. (2009) Response prediction to a multitargeted kinase inhibitor in cancer cell lines and xenograft tumors using high-content tyrosine peptide arrays with a kinetic readout. Mol Cancer Ther, 8, 1846-1855. 81 Roorda, B.D., Ter Elst, A., Diks, S.H., Meeuwsen-de Boer, T.G., Kamps, W.A. and de Bont, E.S. (2009) PTK787/ZK 222584 inhibits tumor growth promoting mesenchymal stem cells: kinase activity profiling as powerful tool in functional studies. Cancer Biol Ther, 8, 1239-1248. 82 Olaussen, K.A., Commo, F., Tailler, M., Lacroix, L., Vitale, I., Raza, S.Q., Richon, C., Dessen, P., Lazar, V., Soria, J.C. et al. (2009) Synergistic proapoptotic effects of the two tyrosine kinase inhibitors pazopanib and lapatinib on multiple carcinoma cell lines. Oncogene, 28, 42494260. 83 Parikh, K., Poppema, S., Peppelenbosch, M.P. and Visser, L. (2009) Extracellular ligationdependent CD45RB enzymatic activity negatively regulates lipid raft signal transduction. Blood, 113, 594-603. 84 Ghosh, G., Yan, X., Lee, A.G., Kron, S.J. and Palecek, S.P. (2010) Quantifying the sensitivities of EGF receptor (EGFR) tyrosine kinase inhibitors in drug resistant non-small cell lung cancer (NSCLC) cells using hydrogel-based peptide array. Biosens Bioelectron, 26, 424-431. 85 Schmerwitz, U.K., Sass, G., Khandoga, A.G., Joore, J., Mayer, B.A., Berberich, N., Totzke, F., Krombach, F., Tiegs, G., Zahler, S. et al. (2011) Flavopiridol Protects Against Inflammation by Attenuating Leukocyte-Endothelial Interaction via Inhibition of Cyclin-Dependent Kinase 9. Arterioscler Thromb Vasc Biol, 31, 280-288. 86 Tegge, W., Frank, R., Hofmann, F. and Dostmann, W.R. (1995) Determination of cyclic nucleotide-dependent protein kinase substrate specificity by the use of peptide libraries on cellulose paper. Biochemistry, 34, 10569-10577. 87 Toomik, R., Edlund, M., Ek, P., Obrink, B. and Engstrom, L. (1996) Simultaneously synthesized peptides on continuous cellulose membranes as substrates for protein kinases. Pept Res, 9, 6-11. 88 Tegge, W.J. and Frank, R. (1998) Analysis of protein kinase substrate specificity by the use of peptide libraries on cellulose paper (SPOT-method). Methods Mol Biol, 87, 99-106. 89 MacBeath, G. and Schreiber, S.L. (2000) Printing proteins as microarrays for high-throughput function determination. Science, 289, 1760-1763. 90 Falsey, J.R., Renil, M., Park, S., Li, S. and Lam, K.S. (2001) Peptide and small molecule microarray for high throughput cell adhesion and functional assays. Bioconjug Chem, 12, 346-353. 91 Lesaicherre, M.L., Uttamchandani, M., Chen, G.Y. and Yao, S.Q. (2002) Antibody-based fluorescence detection of kinase activity on a peptide array. Bioorg Med Chem Lett, 12, 2085-2088. 92 Espanel, X., Walchli, S., Ruckle, T., Harrenga, A., Huguenin-Reggiani, M. and Hooft van Huijsduijnen, R. (2003) Mapping of synergistic components of weakly interacting proteinprotein motifs using arrays of paired peptides. J Biol Chem, 278, 15162-15167. 93 Martin, K., Steinberg, T.H., Goodman, T., Schulenberg, B., Kilgore, J.A., Gee, K.R., Beechem, J.M. and Patton, W.F. (2003) Strategies and solid-phase formats for the analysis of protein and peptide phosphorylation employing a novel fluorescent phosphorylation sensor dye. Comb Chem High Throughput Screen, 6, 331-339. 94 Martin, K., Steinberg, T.H., Cooley, L.A., Gee, K.R., Beechem, J.M. and Patton, W.F. (2003) Quantitative analysis of protein phosphorylation status and protein kinase activity on microarrays using a novel fluorescent phosphorylation sensor dye. Proteomics, 3, 12441255. 95 Schutkowski, M., Reimer, U., Panse, S., Dong, L., Lizcano, J.M., Alessi, D.R. and SchneiderMergener, J. (2004) High-content peptide microarrays for deciphering kinase specificity and biology. Angew Chem Int Ed Engl, 43, 2671-2674. 96 Min, D.H., Su, J. and Mrksich, M. (2004) Profiling kinase activities by using a peptide chip and mass spectrometry. Angew Chem Int Ed Engl, 43, 5973-5977. 97 Wang, Z., Lee, J., Cossins, A.R. and Brust, M. (2005) Microarray-based detection of protein binding and functionality by gold nanoparticle probes. Anal Chem, 77, 5770-5774. 98 Su, J., Bringer, M.R., Ismagilov, R.F. and Mrksich, M. (2005) Combining microfluidic networks and peptide arrays for multi-enzyme assays. J Am Chem Soc, 127, 7280-7281. 99 Inamori, K., Kyo, M., Nishiya, Y., Inoue, Y., Sonoda, T., Kinoshita, E., Koike, T. and Katayama, Y. (2005) Detection and quantification of on-chip phosphorylated peptides by surface plasmon resonance imaging techniques using a phosphate capture molecule. Anal Chem, 77, 3979-3985. 100 Rupcich, N., Green, J.R. and Brennan, J.D. (2005) Nanovolume kinase inhibition assay using a sol-gel-derived multicomponent microarray. Anal Chem, 77, 8013-8019. 101 Horiuchi, K.Y., Wang, Y., Diamond, S.L. and Ma, H. (2006) Microarrays for the functional analysis of the chemical-kinase interactome. J Biomol Screen, 11, 48-56. 102 Horiuchi, K.Y., Wang, Y., Diamond, S.L. and Ma, H. (2006) Microarrays for the functional analysis of the chemical-kinase interactome. J Biomol Screen, 11, 48-56. 103 Sun, L., Liu, D. and Wang, Z. (2007) Microarray-based kinase inhibition assay by gold nanoparticle probes. Anal Chem, 79, 773-777. 104 Shults, M.D., Kozlov, I.A., Nelson, N., Kermani, B.G., Melnyk, P.C., Shevchenko, V., Srinivasan, A., Musmacker, J., Hachmann, J.P., Barker, D.L. et al. (2007) A multiplexed protein kinase assay. Chembiochem, 8, 933-942. 105 Akita, S., Umezawa, N., Kato, N. and Higuchi, T. (2008) Array-based fluorescence assay for serine/threonine kinases using specific chemical reaction. Bioorg Med Chem, 16, 7788-7794. 106 Thiele, A., Zerweck, J., Weiwad, M., Fischer, G. and Schutkowski, M. (2009) High-density Peptide microarrays for reliable identification of phosphorylation sites and upstream kinases. Methods Mol Biol, 570, 203-219. 107 Parikh, K., Peppelenbosch, M.P. and Ritsema, T. (2009) Kinome profiling using peptide arrays in eukaryotic cells. Methods Mol Biol, 527, 269-280, x. 108 Hilhorst, R., Houkes, L., van den Berg, A. and Ruijtenbeek, R. (2009) Peptide microarrays for detailed, high-throughput substrate identification, kinetic characterization, and inhibition studies on protein kinase A. Anal Biochem, 387, 150-161. 109 Poot, A.J., van Ameijde, J., Slijper, M., van den Berg, A., Hilhorst, R., Ruijtenbeek, R., Rijkers, D.T. and Liskamp, R.M. (2009) Development of selective bisubstrate-based inhibitors against protein kinase C (PKC) isozymes by using dynamic peptide microarrays. Chembiochem, 10, 2042-2051. 110 Li, T., Liu, D. and Wang, Z. (2009) Microarray-based Raman spectroscopic assay for kinase inhibition by gold nanoparticle probes. Biosens Bioelectron, 24, 3335-3339. 111 Lee, D.W., Kim, H.J., Choi, C.H., Shin, J.H. and Kim, E.K. (2010) Development of a Protein Chip to Measure PKCbeta Activity. Appl Biochem Biotechnol. 112 Han, X. and Katayama, Y. (2010) A Peptide microarray for detecting protein kinase activity in cell lysates. Methods Mol Biol, 669, 183-194. 113 Thiele, A., Weiwad, M., Zerweck, J., Fischer, G. and Schutkowski, M. (2010) High density peptide microarrays for proteome-wide fingerprinting of kinase activities in cell lysates. Methods Mol Biol, 669, 173-181. 114 Lesaicherre, M.L., Uttamchandani, M., Chen, G.Y. and Yao, S.Q. (2002) Developing sitespecific immobilization strategies of peptides in a microarray. Bioorg Med Chem Lett, 12, 2079-2083. 115 Reimer, U., Reineke, U. and Schneider-Mergener, J. (2002) Peptide arrays: from macro to micro. Curr Opin Biotechnol, 13, 315-320. 116 Uttamchandani, M., Chen, G.Y., Lesaicherre, M.L. and Yao, S.Q. (2004) Site-specific peptide immobilization strategies for the rapid detection of kinase activity on microarrays. Methods Mol Biol, 264, 191-204. 117 Rodriguez, M., Li, S.S., Harper, J.W. and Songyang, Z. (2004) An oriented peptide array library (OPAL) strategy to study protein-protein interactions. J Biol Chem, 279, 8802-8807. 118 Lee, S.J. and Lee, S.Y. (2004) Microarrays of peptides elevated on the protein layer for efficient protein kinase assay. Anal Biochem, 330, 311-316. 119 Kimura, N., Okegawa, T., Yamazaki, K. and Matsuoka, K. (2007) Site-specific, covalent attachment of poly(dT)-modified peptides to solid surfaces for microarrays. Bioconjug Chem, 18, 1778-1785. 120 Kerman, K., Chikae, M., Yamamura, S. and Tamiya, E. (2007) Gold nanoparticle-based electrochemical detection of protein phosphorylation. Anal Chim Acta, 588, 26-33. 121 Inamori, K., Kyo, M., Matsukawa, K., Inoue, Y., Sonoda, T., Tatematsu, K., Tanizawa, K., Mori, T. and Katayama, Y. (2008) Optimal surface chemistry for peptide immobilization in on-chip phosphorylation analysis. Anal Chem, 80, 643-650. 122 Wong, E.Y. and Diamond, S.L. (2008) Enzyme microarrays assembled by acoustic dispensing technology. Anal Biochem, 381, 101-106. 123 Espanel, X., Huguenin-Reggiani, M. and Hooft van Huijsduijnen, R. (2002) The SPOT technique as a tool for studying protein tyrosine phosphatase substrate specificities. Protein Sci, 11, 2326-2334. 124 Pasquali, C., Curchod, M.L., Walchli, S., Espanel, X., Guerrier, M., Arigoni, F., Strous, G. and Van Huijsduijnen, R.H. (2003) Identification of protein tyrosine phosphatases with specificity for the ligand-activated growth hormone receptor. Mol Endocrinol, 17, 2228-2239. 125 Espanel, X. and Hooft van Huijsduijnen, R. (2005) Applying the SPOT peptide synthesis procedure to the study of protein tyrosine phosphatase substrate specificity: probing for the heavenly match in vitro. Methods, 35, 64-72. 126 Kohn, M., Gutierrez-Rodriguez, M., Jonkheijm, P., Wetzel, S., Wacker, R., Schroeder, H., Prinz, H., Niemeyer, C.M., Breinbauer, R., Szedlacsek, S.E. et al. (2007) A microarray strategy for mapping the substrate specificity of protein tyrosine phosphatase. Angew Chem Int Ed Engl, 46, 7700-7703. 127 Sun, H., Lu, C.H., Uttamchandani, M., Xia, Y., Liou, Y.C. and Yao, S.Q. (2008) Peptide microarray for high-throughput determination of phosphatase specificity and biology. Angew Chem Int Ed Engl, 47, 1698-1702. 128 Govindaraju, T., Jonkheijm, P., Gogolin, L., Schroeder, H., Becker, C.F., Niemeyer, C.M. and Waldmann, H. (2008) Surface immobilization of biomolecules by click sulfonamide reaction. Chem Commun (Camb), 3723-3725. 129 Sacco, F., Tinti, M., Palma, A., Ferrari, E., Nardozza, A.P., Hooft van Huijsduijnen, R., Takahashi, T., Castagnoli, L. and Cesareni, G. (2009) Tumor suppressor density-enhanced phosphatase-1 (DEP-1) inhibits the RAS pathway by direct dephosphorylation of ERK1/2 kinases. J Biol Chem. 130 Sun, H., Tan, L.P., Gao, L. and Yao, S.Q. (2009) High-throughput screening of catalytically inactive mutants of protein tyrosine phosphatases (PTPs) in a phosphopeptide microarray. Chem Commun (Camb), 677-679. 131 Gao, L., Sun, H. and Yao, S.Q. (2010) Activity-based high-throughput determination of PTPs substrate specificity using a phosphopeptide microarray. Biopolymers, 94, 810-819. 132 Ferrari, E., Tinti, M., Costa, S., Corallino, S., Nardozza, A.P., Chatr-Aryamontri, A., Ceol, A., Cesareni, G. and Castagnoli, L. (2010) Identification of new substrates of the protein tyrosine phosphatase PTP1B by bayesian integration of proteome evidence. J Biol Chem. 133 Rathert, P., Dhayalan, A., Murakami, M., Zhang, X., Tamas, R., Jurkowska, R., Komatsu, Y., Shinkai, Y., Cheng, X. and Jeltsch, A. (2008) Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol, 4, 344-346. 134 Rathert, P., Zhang, X., Freund, C., Cheng, X. and Jeltsch, A. (2008) Analysis of the substrate specificity of the dim-5 histone lysine methyltransferase using Peptide arrays. Chem Biol, 15, 5-11. 135 Dhayalan, A., Kudithipudi, S., Rathert, P. and Jeltsch, A. (2011) Specificity Analysis-Based Identification of New Methylation Targets of the SET7/9 Protein Lysine Methyltransferase. Chem Biol, 18, 111-120. 136 Lu, P.J., Zhou, X.Z., Shen, M. and Lu, K.P. (1999) Function of WW domains as phosphoserineor phosphothreonine-binding modules. Science, 283, 1325-1328. 137 Patzelt, H., Rudiger, S., Brehmer, D., Kramer, G., Vorderwulbecke, S., Schaffitzel, E., Waitz, A., Hesterkamp, T., Dong, L., Schneider-Mergener, J. et al. (2001) Binding specificity of Escherichia coli trigger factor. Proc Natl Acad Sci U S A, 98, 14244-14249. 138 Deuerling, E., Patzelt, H., Vorderwulbecke, S., Rauch, T., Kramer, G., Schaffitzel, E., Mogk, A., Schulze-Specking, A., Langen, H. and Bukau, B. (2003) Trigger Factor and DnaK possess overlapping substrate pools and binding specificities. Mol Microbiol, 47, 1317-1328. 139 Wildemann, D., Erdmann, F., Alvarez, B.H., Stoller, G., Zhou, X.Z., Fanghanel, J., Schutkowski, M., Lu, K.P. and Fischer, G. (2006) Nanomolar inhibitors of the peptidyl prolyl cis/trans isomerase Pin1 from combinatorial peptide libraries. J Med Chem, 49, 2147-2150. 140 Lee, J. and Bedford, M.T. (2002) PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep, 3, 268-273. 141 Min, D.H., Yeo, W.S. and Mrksich, M. (2004) A method for connecting solution-phase enzyme activity assays with immobilized format analysis by mass spectrometry. Anal Chem, 76, 3923-3929. 142 Schwamborn, K., Knipscheer, P., van Dijk, E., van Dijk, W.J., Sixma, T.K., Meloen, R.H. and Langedijk, J.P. (2008) SUMO assay with peptide arrays on solid support: insights into SUMO target sites. J Biochem, 144, 39-49. 143 Li, X., Vadrevu, S., Dunlop, A.J., Day, J., Advant, N., Troeger, J., Klussmann, E., Jaffray, E.G., Hay, R.T., Adams, D.R. et al. (2010) Selective SUMO modification of cAMP-specific phosphodiesterase-4D5 (PDE4D5) regulates the functional consequences of phosphorylation by PKA and ERK. Biochem J. 144 Li, X., Baillie, G.S. and Houslay, M.D. (2009) Mdm2 directs the ubiquitination of betaarrestin-sequestered cAMP phosphodiesterase-4D5. J Biol Chem, 284, 16170-16182. 145 Laurent, N., Voglmeir, J., Wright, A., Blackburn, J., Pham, N.T., Wong, S.C., Gaskell, S.J. and Flitsch, S.L. (2008) Enzymatic glycosylation of peptide arrays on gold surfaces. Chembiochem, 9, 883-887. 146 Blixt, O., Clo, E., Nudelman, A.S., Sorensen, K.K., Clausen, T., Wandall, H.H., Livingston, P.O., Clausen, H. and Jensen, K.J. (2010) A high-throughput O-glycopeptide discovery platform for seromic profiling. J Proteome Res, 9, 5250-5261. 147 Steentoft, C., Schjoldager, K.T., Clo, E., Mandel, U., Levery, S.B., Pedersen, J.W., Jensen, K., Blixt, O. and Clausen, H. (2010) Characterization of an immunodominant cancer-specific Oglycopeptide epitope in murine podoplanin (OTS8). Glycoconj J, 27, 571-582. 148 von Olleschik-Elbheim, L., el Bayâ, A. and Schmidt, M.A. (1997) ADP-Ribosylation in Animal Tissue (Ed. Haag, Koch-Nolte), 87. 149 Clemente, S., Franco, L. and Lopez-Rodas, G. (2001) Distinct site specificity of two pea histone deacetylase complexes. Biochemistry, 40, 10671-10676. 150 Gurard-Levin, Z.A. and Mrksich, M. (2008) The activity of HDAC8 depends on local and distal sequences of its peptide substrates. Biochemistry, 47, 6242-6250. 151 Gurard-Levin, Z.A., Kim, J. and Mrksich, M. (2009) Combining mass spectrometry and peptide arrays to profile the specificities of histone deacetylases. Chembiochem, 10, 21592161. 152 Fahie, K., Hu, P., Swatkoski, S., Cotter, R.J., Zhang, Y. and Wolberger, C. (2009) Side chain specificity of ADP-ribosylation by a sirtuin. FEBS J, 276, 7159-7176. 153 Smith, B.C., Settles, B., Hallows, W.C., Craven, M.W. and Denu, J.M. (2010) SIRT3 Substrate Specificity Determined by Peptide Arrays and Machine Learning. ACS Chem Biol. 154 Gurard-Levin, Z.A., Kilian, K.A., Kim, J., Bahr, K. and Mrksich, M. (2010) Peptide arrays identify isoform-selective substrates for profiling endogenous lysine deacetylase activity. ACS Chem Biol, 5, 863-873. 155 Hallows, W.C., Yu, W., Smith, B.C., Devires, M.K., Ellinger, J.J., Someya, S., Shortreed, M.R., Prolla, T., Markley, J.L., Smith, L.M. et al. (2011) Sirt3 Promotes the Urea Cycle and Fatty Acid Oxidation during Dietary Restriction. Mol Cell, 41, 139-149. 156 Duan, Y. and Laursen, R.A. (1994) Protease substrate specificity mapping using membranebound peptides. Anal Biochem, 216, 431-438. 157 Hilpert, K., Hansen, G., Wessner, H., Schneider-Mergener, J. and Hohne, W. (2000) Characterizing and optimizing protease/peptide inhibitor interactions, a new application for spot synthesis. J Biochem, 128, 1051-1057. 158 Dekker, N., Cox, R.C., Kramer, R.A. and Egmond, M.R. (2001) Substrate specificity of the integral membrane protease OmpT determined by spatially addressed peptide libraries. Biochemistry, 40, 1694-1701. 159 Winssinger, N., Harris, J.L., Backes, B.J. and Schultz, P.G. (2001) Angew. Chem., 113, 32543258. 160 Kaup, M., Dassler, K., Reineke, U., Weise, C., Tauber, R. and Fuchs, H. (2002) Processing of the human transferrin receptor at distinct positions within the stalk region by neutrophil elastase and cathepsin G. Biol Chem, 383, 1011-1020. 161 Winssinger, N., Ficarro, S., Schultz, P.G. and Harris, J.L. (2002) Profiling protein function with small molecule microarrays. Proc Natl Acad Sci U S A, 99, 11139-11144. 162 Jones, C.H., Dexter, P., Evans, A.K., Liu, C., Hultgren, S.J. and Hruby, D.E. (2002) Escherichia coli DegP protease cleaves between paired hydrophobic residues in a natural substrate: the PapA pilin. J Bacteriol, 184, 5762-5771. 163 Salisbury, C.M., Maly, D.J. and Ellman, J.A. (2002) Peptide microarrays for the determination of protease substrate specificity. J Am Chem Soc, 124, 14868-14870. 164 Gosalia, D.N. and Diamond, S.L. (2003) Printing chemical libraries on microarrays for fluid phase nanoliter reactions. Proc Natl Acad Sci U S A, 100, 8721-8726. 165 Winssinger, N., Damoiseaux, R., Tully, D.C., Geierstanger, B.H., Burdick, K. and Harris, J.L. (2004) PNA-encoded protease substrate microarrays. Chem Biol, 11, 1351-1360. 166 Kiyonaka, S., Sada, K., Yoshimura, I., Shinkai, S., Kato, N. and Hamachi, I. (2004) Semi-wet peptide/protein array using supramolecular hydrogel. Nat Mater, 3, 58-64. 167 Janssen, S., Jakobsen, C.M., Rosen, D.M., Ricklis, R.M., Reineke, U., Christensen, S.B., Lilja, H. and Denmeade, S.R. (2004) Screening a combinatorial peptide library to develop a human glandular kallikrein 2-activated prodrug as targeted therapy for prostate cancer. Mol Cancer Ther, 3, 1439-1450. 168 Wegner, G.J., Wark, A.W., Lee, H.J., Codner, E., Saeki, T., Fang, S. and Corn, R.M. (2004) Realtime surface plasmon resonance imaging measurements for the multiplexed determination of protein adsorption/desorption kinetics and surface enzymatic reactions on peptide microarrays. Anal Chem, 76, 5677-5684. 169 Gosalia, D.N., Salisbury, C.M., Ellman, J.A. and Diamond, S.L. (2005) High throughput substrate specificity profiling of serine and cysteine proteases using solution-phase fluorogenic peptide microarrays. Mol Cell Proteomics, 4, 626-636. 170 Gosalia, D.N., Salisbury, C.M., Maly, D.J., Ellman, J.A. and Diamond, S.L. (2005) Profiling serine protease substrate specificity with solution phase fluorogenic peptide microarrays. Proteomics, 5, 1292-1298. 171 Gosalia, D.N., Denney, W.S., Salisbury, C.M., Ellman, J.A. and Diamond, S.L. (2006) Functional phenotyping of human plasma using a 361-fluorogenic substrate biosensing microarray. Biotechnol Bioeng, 94, 1099-1110. 172 Naus, S., Reipschlager, S., Wildeboer, D., Lichtenthaler, S.F., Mitterreiter, S., Guan, Z., Moss, M.L. and Bartsch, J.W. (2006) Identification of candidate substrates for ectodomain shedding by the metalloprotease-disintegrin ADAM8. Biol Chem, 387, 337-346. 173 Diaz-Mochon, J.J., Bialy, L. and Bradley, M. (2006) Dual colour, microarray-based, analysis of 10,000 protease substrates. Chem Commun (Camb), 3984-3986. 174 Han, A., Sonoda, T., Kang, J.H., Murata, M., T, N.I. and Katayam, Y. (2006) Development of a fluorescence peptide chip for the detection of caspase activity. Comb Chem High Throughput Screen, 9, 21-25. 175 Su, J., Rajapaksha, T.W., Peter, M.E. and Mrksich, M. (2006) Assays of endogenous caspase activities: a comparison of mass spectrometry and fluorescence formats. Anal Chem, 78, 4945-4951. 176 Kim, D.H., Shin, D.S. and Lee, Y.S. (2007) Spot arrays on modified glass surfaces for efficient SPOT synthesis and on-chip bioassay of peptides. J Pept Sci, 13, 625-633. 177 Inoue, Y., Mori, T., Yamanouchi, G., Han, X., Sonoda, T., Niidome, T. and Katayama, Y. (2008) Surface plasmon resonance imaging measurements of caspase reactions on peptide microarrays. Anal Biochem, 375, 147-149. 178 Kozlov, I.A., Melnyk, P.C., Hachmann, J.P., Srinivasan, A., Shults, M., Zhao, C., Musmacker, J., Nelson, N., Barker, D.L. and Lebl, M. (2008) A high-complexity, multiplexed solution-phase assay for profiling protease activity on microarrays. Comb Chem High Throughput Screen, 11, 24-35. 179 Sapsford, K.E., Sun, S., Francis, J., Sharma, S., Kostov, Y. and Rasooly, A. (2008) A fluorescence detection platform using spatial electroluminescent excitation for measuring botulinum neurotoxin A activity. Biosens Bioelectron, 24, 618-625. 180 Cedzich, A., Huttenlocher, F., Kuhn, B.M., Pfannstiel, J., Gabler, L., Stintzi, A. and Schaller, A. (2009) The protease-associated domain and C-terminal extension are required for zymogen processing, sorting within the secretory pathway, and activity of tomato subtilase 3 (SlSBT3). J Biol Chem, 284, 14068-14078. 181 Li, J. and Yao, S.Q. (2009) "Singapore Green": a new fluorescent dye for microarray and bioimaging applications. Org Lett, 11, 405-408. 182 Bovenschen, N., Quadir, R., van den Berg, A.L., Brenkman, A.B., Vandenberghe, I., Devreese, B., Joore, J. and Kummer, J.A. (2009) Granzyme K displays highly restricted substrate specificity that only partially overlaps with granzyme A. J Biol Chem, 284, 3504-3512. 183 Collet, B.Y., Nagashima, T., Yu, M.S. and Pohl, N.L. (2009) Fluorous-based Peptide Microarrays for Protease Screening. J Fluor Chem, 130, 1042-1048. 184 Zhu, Q., Uttamchandani, M., Li, D., Lesaicherre, M.L. and Yao, S.Q. (2003) Enzymatic profiling system in a small-molecule microarray. Org Lett, 5, 1257-1260. 185 Fu, J., Cai, K., Johnston, S.A. and Woodbury, N.W. (2010) Exploring peptide space for enzyme modulators. J Am Chem Soc, 132, 6419-6424.