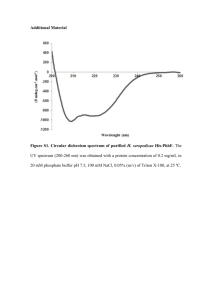
bit25658-sup-0001-SupFigLeg-S1
advertisement

1 SUPPLEMENTARY FIGURE 1. SDS-PAGE analysis of purified Hyp-complexes and 2 maturases. Proteins and complexes were purified from E. coli BL21Star™ (DE3) cells 3 containing plasmid constructs listed in Table I. Gene expression was performed as described 4 in the methods section. Proteins were separated by SDS-PAGE and the gels were stained by 5 Coomassie Brilliant Blue. Purified complexes (acrylamide concentration of the gels): A) 6 HypC1-StrepII/HoxH-complex (12%/18%); B) HypC1-StrepII/HypD1-complex (12%/18%); 7 C) HypC1-StrepII/HypD1/HypE1-complex (12%/18%); D) HypA2/StrepII-HypB2-complex 8 (12%/18%); E) HypC2-StrepII/HoxH-complex (12%/18%); F) HypC2-StrepII/HypD2- 9 complex (12%/18%); G) HypC2-StrepII/HypD2/HypE2-complex (18%); H) HypA3/StrepII- 10 HypB3-complex (18%); I) HypC1-StrepII/HypD2-complex (18%). 25 µg cell-free extract or 11 4 µg (A, B, D–H) to 6 µg (C, I) of purified complexes were applied per lane. Fragments of 12 gels were composed to highlight relevant lanes. Legend: M = Marker/Protein ladder; CFE = 13 Cell-free soluble extract; E = Elution fraction; C = contaminant. 14 15 SUPPLEMENTARY FIGURE 2. Sequence alignment of related HoxW-proteins. The 16 conserved nickel binding motif present in all HoxW protein sequences is highlighted in green. 17 Cysteines with the capacity to coordinate a putative [4Fe-4S]-cluster are highlighted in red, as 18 are the cysteines in the other sequences at the same position. Notably, two homologues (from 19 Cn and Mv) contain a complete CxxCxxC motif. A fourth cysteine positioned outside the 20 CxxCxxC-motif is highlighted in yellow (in HoxW: residue no. 49). It must be stressed that a 21 putative fourth coordinating residue could be taken by a number of amino acids other than 22 cysteine. Legend: Av = Azotobacter vinelandii HoxW; Gh = Grimontia hollisae HoxW; Mv = 23 Mycobacterium vanbaalenii HoxW; Ro = Rhodococcus opacus HoxW; Cn = Cupriavidus 24 necator HoxW; Rm = Ralstonia metallidurans HoxW; Nitr = Nitrosospira sp. APG3 HoxW; 25 Mc = Methylococcus capsulatus HoxW; Syn = Synechococcus sp. PCC 7002 HoxW; Lm = 26 Lyngbya majuscula HoxW. 27 28 SUPPLEMENTARY FIGURE 3. Analysis of the purified HypCDE-complex. The complex 29 was purified from E. coli BL21Star™ (pE-[3’]hypC1[wt]hypD1[wt]hypE1) cells subjected to 30 autoinduction according to the procedure described in the methods section of the article. A) 31 Purification of the ternary complex by affinity chromatography, visually followed by SDS- 32 PAGE on 12/18%-gels stained by Coomassie Brilliant Blue. The two weak bands between 33 30–40 kDa are contaminants associated with HypE1, which were also present upon individual 34 purification of StrepII-HypE1. 25 µg of cell-free extract and 6 µg of purified complex were 35 applied. Legend: M = SDS-PAGE marker/protein ladder; CFE = cell-free soluble extract; E = 36 elution fraction. B) Native PAGE (N-PAGE) and Western-Blotting (WB) of the purified 37 complex. N-PAGE was performed on 4–15% gradient gels. 6 µg of purified complex were 38 applied for the Coomassie-stained gel, whereas 0.5 µg were used for subsequent WB and 39 chemiluminescence detection. Legend: M = N-PAGE marker/protein ladder; E = elution 40 fraction (Coomassie-stained); B = elution fraction (WB-visualization). Fragments of gels in 41 A) and B) were composed to highlight relevant lanes. C) Molecular mass determination of 42 HypC1D1E1-species by analytical size-exclusion chromatography. Molecular masses were 43 plotted on a logarithmic scale. Standards and samples were analyzed on a Superdex 200 HR 44 10/300 gel filtration column. Buffers and standards used are described in the methods section 45 of the article. A linear fit with the formula K = –0.14·ln(Mr) + 1.22 (R2 = 0.982) was used for 46 molecular mass determination. The calculated masses of the two distinguished species were 47 91 kDa (K = 0.601), corresponding to a monomeric HypC1D1E1-complex and 179 kDa (K = 48 0.508), which corresponds to a (HypC1D1E1)2-complex. Legend: K = partition coefficient; 49 Mr = relative molecular mass [kDa].