Abstract - Department of Chemistry
advertisement
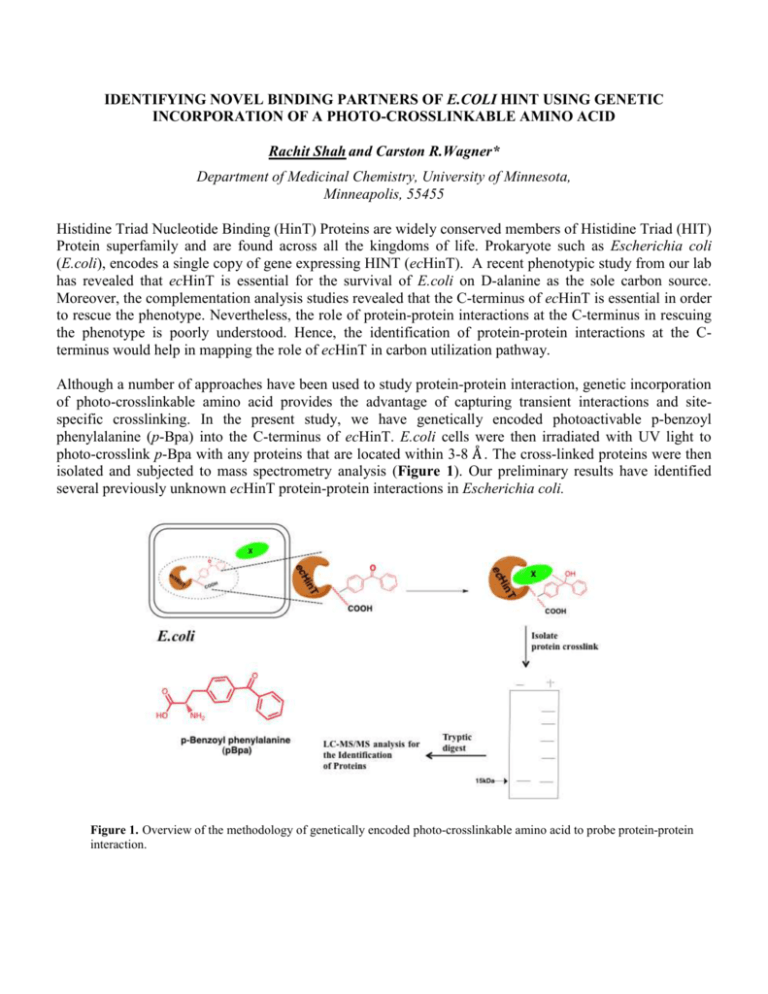
IDENTIFYING NOVEL BINDING PARTNERS OF E.COLI HINT USING GENETIC INCORPORATION OF A PHOTO-CROSSLINKABLE AMINO ACID Rachit Shah and Carston R.Wagner* Department of Medicinal Chemistry, University of Minnesota, Minneapolis, 55455 Histidine Triad Nucleotide Binding (HinT) Proteins are widely conserved members of Histidine Triad (HIT) Protein superfamily and are found across all the kingdoms of life. Prokaryote such as Escherichia coli (E.coli), encodes a single copy of gene expressing HINT (ecHinT). A recent phenotypic study from our lab has revealed that ecHinT is essential for the survival of E.coli on D-alanine as the sole carbon source. Moreover, the complementation analysis studies revealed that the C-terminus of ecHinT is essential in order to rescue the phenotype. Nevertheless, the role of protein-protein interactions at the C-terminus in rescuing the phenotype is poorly understood. Hence, the identification of protein-protein interactions at the Cterminus would help in mapping the role of ecHinT in carbon utilization pathway. Although a number of approaches have been used to study protein-protein interaction, genetic incorporation of photo-crosslinkable amino acid provides the advantage of capturing transient interactions and sitespecific crosslinking. In the present study, we have genetically encoded photoactivable p-benzoyl phenylalanine (p-Bpa) into the C-terminus of ecHinT. E.coli cells were then irradiated with UV light to photo-crosslink p-Bpa with any proteins that are located within 3-8 Å . The cross-linked proteins were then isolated and subjected to mass spectrometry analysis (Figure 1). Our preliminary results have identified several previously unknown ecHinT protein-protein interactions in Escherichia coli. Figure 1. Overview of the methodology of genetically encoded photo-crosslinkable amino acid to probe protein-protein interaction.