Table S1. List of strains and plasmids used in this study. Strain
advertisement

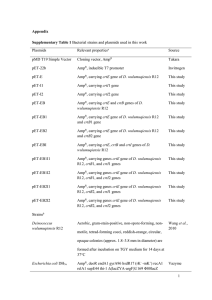
Table S1. List of strains and plasmids used in this study. Phenotype, genotype and/or descriptiona Ref/Source TOP10 F– mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL (Strr) endA1 nupG Invitrogen GM2163 dam-13::Tn 9 dcm6 hsdR2 leuB6 his-4 thi-1 ara-14 lacY1 galK2 galT22 xyl-5 mtl-1 rpsL136 tonA31 tsx-78 supE44 McrA- McrB New England Biolabs Rosetta (DE3) F- ompT hsdSB(rB- mB-) gal dcm (DE3) pRARE (Cmr) Novagen H26 DS70 pyrE2 (Allers et al, 2004) GZ108 H26 ΔpanB (Zhou et al, 2008) HM1052 H26 ΔubaA (Miranda et al, 2011) Plasmidsb pET24b Cmr, Kmr, expression vector for E. coli (DE3) strains Strain, plasmid E. coli Hfx. volcanii (Humbard et al, 2009) pJAM2001 Ampr Nvr; C-terminal StrepII expression vector for Hfx. volcanii strains Ampr Nvr; PAN-A/1-StrepII pJAM1106 Ampr Nvr; Flag-His-SAMP1 in pJAM202c This study pJAM1119 Ampr Nvr; MoaE-StrepII (Hepowit et al, 2012) pJAM1796 Ampr Nvr; Flag-SAMP1-MoaE-M1A-StrepII (Hepowit et al, 2012) pJAM1131 Cmr, Kmr, Flag-His-SAMP1 in pET24b This study pJAM809 This study a Abbreviations: Ampr, ampicillin resistance; Nvr, novobiocin resistance ; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; -StrepII, C-terminal StrepII tag fusion protein; His-, N-terminal poly-His6 tag fusion protein; Flag-, N-terminal Flag tag fusion protein; Flag-His-, N-terminal tandem affinity Flag and poly-His6 tagged protein. b To generate pJAM2001, PCR was used with primers 1/2 to isolate an Hfx. volcanii genomic fragment carrying the panA/1 gene that was ligated into the NdeI to KpnI sites of plasmid pJAM809. To generate pJAM1106, PCR was used with primers 5/6 to generate an Hfx. volcanii genomic fragment carrying the samp1 gene that was ligated into the KpnI to BlpI sites of pJAM939. Plasmid pJAM1131 was derived by NdeI to BlpI ligation of the Flag-His6-SAMP1 coding region of pJAM1106 into plasmid vector pET24b. PCRs were with Hfx. volcanii H26 or DS70 genomic DNA as the template using primers listed in Table S2 and Phusion® High-Fidelity DNA Polymerase according to Manufacturer’s protocol (New England Biolabs). DNA fragments were treated with Antarctic Phosphatase and/or extracted from gels after separation by 0.8% (w/v) agarose gel electrophoresis in 1X TAE buffer with MinElute® gel extraction kit (Qiagen) as needed. Plasmid vectors and digested PCR-amplified products were ligated overnight at 16˚C with T4 DNA ligase (New England Biolabs). The fidelity of all plasmids was confirmed by DNA sequencing (ICBR DNA Sequencing Core, University of Florida). 1 Table S2. List of primers used in this study. Primer no. 1 Primer name Oligonucletide Sequence (5’ to 3’)a HVPanA Nde1 FW 5’-CGGCATcatatgATGACCGATACT-3’ 2 HVPanA Kpn1 RV 5’-ATggtacccGCGAACGCGC-3’ 3 Hvo_2619 NdeI up 5’-CTGCCCTcatatgGAGTGGAAGCTGT-3’ 4 Hvo_2619 BlpI down 5’-TTAATgctcagcTCACCCACCGGC-3’ 5 HVO_2619 KpnI-His Up 5’-AAggtaccCACCACCACCACCACCACGAGTGG AAGCTGTTC-3’ 6 HVO_2619 BlpI down 5’-TTAATgctcagcCTAGCCGCCGCTGACCGG-3’ a Lowercase letters indicate restriction enzyme site. Italicized uppercase letters indicate random bases introduced into primer to enhance cleavage of PCR product by restriction enzyme. 2 Table S3. Modified guanidine-HCl buffered conditions used for denaturing and renaturing the halophilic proteins by far western blottinga. Buffer Reagents/ Conditions 8 M Guanidine-HCl (mL) Glycerol (mL) NaCl (g) 500 mM HEPES (mL) 0.5 M EDTA (mL) 10% Tween-20 (mL) Milk powder (g) 1M DTT (µL) ddH2O (mL) Total volume (mL) Time/Temperature 1 150 (6M) 20 1.16 (0.1 M) 8 0.4 2 4 200 21.4 200 30 min/RT Far Western Blotting Steps 2 3 4 74.4 (3 M) 24.8 (1 M) 2.48 (0.1 M) 20 20 20 1.16 (0.1 M) 1.16 (0.1 M) 2.32 (0.2 M) 8 8 8 0.4 0.4 0.4 2 2 2 4 4 4 200 200 200 97 146.6 168.92 200 200 200 30 min/RT 30 min/RT 30 min/4°C a 5 0 (0 M) 20 24 (2 M) 8 0.4 2 4 200 171.4 200 overnight/4°C Protocol was adapted from the far western blotting method of (Wu et al, 2007) for the halophilic (saltloving) proteins of this study. All solutions were freshly prepared prior to use. Membrane-bound proteins were denatured in step 1 and renatured in a stepwise fashion (steps 2 to 5) resulting in a decrease in the concentration of guanidine-HCl from 6 to 0 M and increase the NaCl concentration from 0.1 to 2 M. The final buffer of step 5 was supplemented with 2M NaCl to enhance renaturation of the halophilic proteins. 3 Supplemental References Allers T, Ngo HP, Mevarech M, Lloyd RG (2004) Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl Environ Microbiol 70: 943-953 Hepowit NL, Uthandi S, Miranda HV, Toniutti M, Prunetti L, Olivarez O, De Vera IM, Fanucci GE, Chen S, Maupin-Furlow JA (2012) Archaeal JAB1/MPN/MOV34 metalloenzyme (HvJAMM1) cleaves ubiquitin-like small archaeal modifier proteins (SAMPs) from protein-conjugates. Mol Microbiol 86: 971-987 Humbard M, Zhou G, Maupin-Furlow J (2009) The N-terminal penultimate residue of 20S proteasome alpha 1 influences its N-alpha acetylation and protein levels as well as growth rate and stress responses of Haloferax volcanii. JOURNAL OF BACTERIOLOGY 191: 3794-3803 Miranda HV, Nembhard N, Su D, Hepowit N, Krause DJ, Pritz JR, Phillips C, Söll D, Maupin-Furlow JA (2011) E1- and ubiquitin-like proteins provide a direct link between protein conjugation and sulfur transfer in archaea. Proc Natl Acad Sci U S A 108: 4417-4422 Wu Y, Li Q, Chen XZ (2007) Detecting protein-protein interactions by Far western blotting. Nat Protoc 2: 3278-3284 Zhou G, Kowalczyk D, Humbard M, Rohatgi S, Maupin-Furlow J (2008) Proteasomal components required for cell growth and stress responses in the haloarchaeon Haloferax volcanii. J Bacteriol 190: 8096-8105 4