Appendix 1. Protocol used for PCR amplification and DNA
advertisement

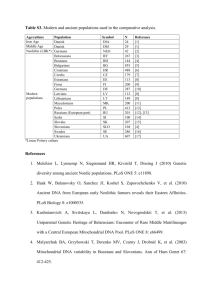
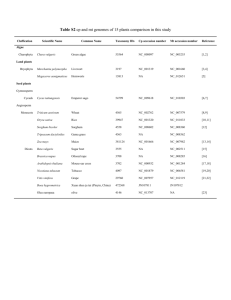
Appendix 1. Protocol used for PCR amplification and DNA sequencing Twenty-four overlapping fragments of the mitochondrial genome were amplified by PCR using primers published in Hassanin et al. (2009), Hassanin et al. (2012) and Hassanin (submitted a,b): U400M1/LPHE; U400M1/12SL200; 12SU1230/12SL2226M1LA; 12SU829/16SL518; 16SU365/16SL1056; 16SU946/N1L64; Uleu/LMet2; N1U840/N2L492; N2U354M2/AsnL; TrpU/C1L705; UTyr/LSer; C1U897M1/C2L15M1; SerU/A8L1; C2U603/C3L45; A6U654M1/GlyLM1; C3U780M3/N4L366M1; UArg/N4L918M1; N4U681/Leu2LM1; Ser2U/N5L652; N5U501/N5L1214; N5U1146M5/N5L154M5; N6RU102/CBL402; GluMA/ProMA; and U844/L482. Amplifications were done in 20 μl using 3 μl of Buffer 10X with MgCl2, 2 μl of dNTP (6.6 mM), 0.12 μl of Taq DNA polymerase (2.5 U, Qiagen, Hilden, Germany) and 0.75 μl of the two primers at 10 μM. The standard PCR conditions were as follows: 4 min at 94°C; 5 cycles of denaturation / annealing / extension with 45s at 94°C, 1 min at 60°C and 1 min at 72°C, followed by 30 cycles of 30s at 94°C, 45s at 55°C, and 1 min at 72°C, followed by 10 min at 72°C. Both strands of PCR products were sequenced using Sanger sequencing on an ABI 3730 automatic sequencer at the Centre National de Séquençage (Genoscope) in Evry (France). The sequences were edited and assembled using Sequencher 5.1 (Gene Codes, Corp., Ann Arbor, MI, USA). References Hassanin A. (2014). The complete mitochondrial genome of the African palm civet, Nandinia binotata, the only representative of the family Nandiniidae (Mammalia, Carnivora). Mitochondrial DNA. Hassanin A. (2014). The complete mitochondrial genome of the Servaline Genet, Genetta servalina, the first representative from the family Viverridae (Mammalia, Carnivora). Mitochondrial DNA. Hassanin A, Delsuc F, Ropiquet A, Hammer C, Jansen van Vuuren B, Matthee C, RuizGarcia M, Catzeflis F, Areskoug V, Nguyen TT, Couloux A. 2012. Pattern and timing of diversification of Cetartiodactyla (Mammalia, Laurasiatheria), as revealed by a comprehensive analysis of mitochondrial genomes. C R Biol. 335:32-50. Hassanin A, Ropiquet A, Couloux A, Cruaud C. 2009. Evolution of the mitochondrial genome in mammals living at high altitude: new insights from a study of the tribe Caprini (Bovidae, Antilopinae). J Mol Evol 68:293-310. Appendix 2. Interfamilial relationships within the order Carnivora, as inferred from a DNA alignment of complete mitochondrial genomes. Our taxonomic sample includes 23 species of the order Carnivora, representing all the families of Feliformia and most families of Caniformia. Four genera, representing different orders of Laurasiatheria, were used to root the tree of Carnivora: Boselaphus (Cetartiodactyla), Ceratotherium (Perissodactyla), Manis (Pholidota), and Rhinolophus (Chiroptera). Accession numbers of the DNA sequences are listed on the tree, after the species names. The complete mitochondrial genomes were aligned by eye using Se-Al v2.0a11 (A. Rambaut, Sequence Alignment Editor Version 2.0 alpha 11, software available at http://evolve.zoo.ox.ac.uk/). All ambiguous regions, i.e., involving ambiguity for the position of the gaps, were excluded from the analyses to avoid erroneous hypotheses of primary homology. For this reason, the control region was not included in the alignment because of the presence of many ambiguous gaps. The reduced alignment consists of 15,080 nt. The phylogenetic tree was constructed on PAUP version 4 (Swofford, 2003) using the Maximum Likelihood (ML) method and the GTR+I+G model selected by selected by the Akaike Information Criterion under jModelTest 2 (Darriba et al., 2012). Bootstrap percentages (BP), which were computed after 100 replicates, are indicated on the branches. References Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9:772. Swofford, DL. 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, Massachusetts.