SystemsLevelForWeb
Systems level insights into alternate methane cycling modes in Lake Washington
Major objectives and specific approaches
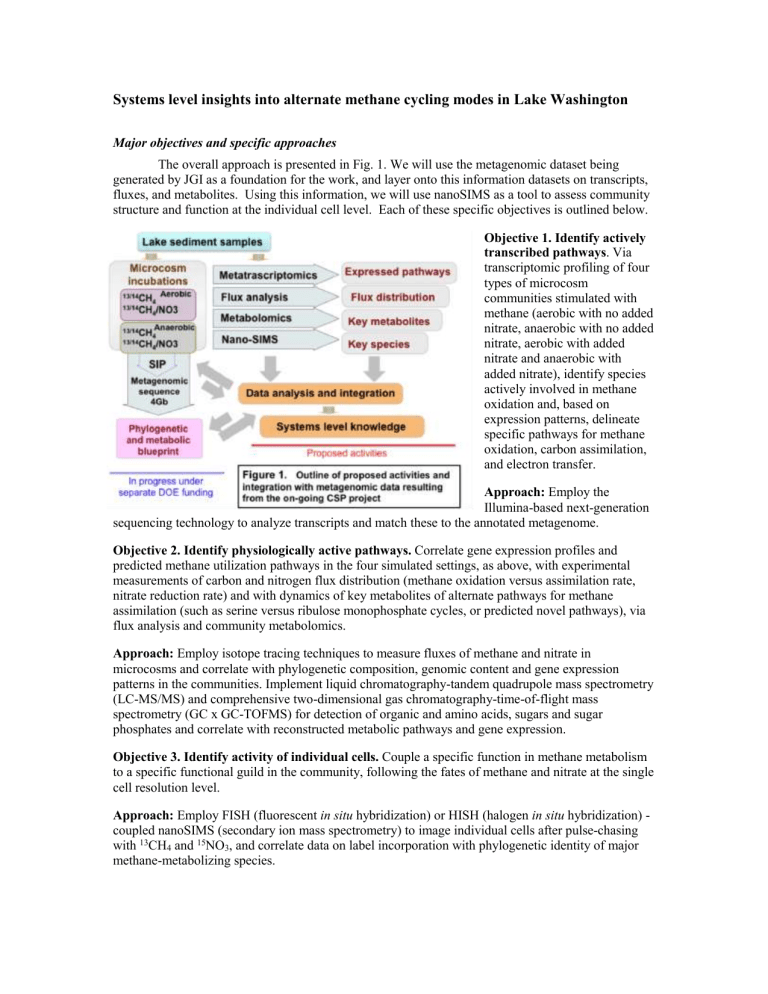
The overall approach is presented in Fig. 1. We will use the metagenomic dataset being generated by JGI as a foundation for the work, and layer onto this information datasets on transcripts, fluxes, and metabolites. Using this information, we will use nanoSIMS as a tool to assess community structure and function at the individual cell level. Each of these specific objectives is outlined below.
Objective 1.
Identify actively transcribed pathways . Via transcriptomic profiling of four types of microcosm communities stimulated with methane (aerobic with no added nitrate, anaerobic with no added nitrate, aerobic with added nitrate and anaerobic with added nitrate), identify species actively involved in methane oxidation and, based on expression patterns, delineate specific pathways for methane oxidation, carbon assimilation, and electron transfer.
Approach: Employ the
Illumina-based next-generation sequencing technology to analyze transcripts and match these to the annotated metagenome.
Objective 2. Identify physiologically active pathways. Correlate gene expression profiles and predicted methane utilization pathways in the four simulated settings, as above, with experimental measurements of carbon and nitrogen flux distribution (methane oxidation versus assimilation rate, nitrate reduction rate) and with dynamics of key metabolites of alternate pathways for methane assimilation (such as serine versus ribulose monophosphate cycles, or predicted novel pathways), via flux analysis and community metabolomics.
Approach: Employ isotope tracing techniques to measure fluxes of methane and nitrate in microcosms and correlate with phylogenetic composition, genomic content and gene expression patterns in the communities. Implement liquid chromatography-tandem quadrupole mass spectrometry
(LC-MS/MS) and comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry (GC x GC-TOFMS) for detection of organic and amino acids, sugars and sugar phosphates and correlate with reconstructed metabolic pathways and gene expression.
Objective 3. Identify activity of individual cells. Couple a specific function in methane metabolism to a specific functional guild in the community, following the fates of methane and nitrate at the single cell resolution level.
Approach: Employ FISH (fluorescent in situ hybridization) or HISH (halogen in situ hybridization) - coupled nanoSIMS (secondary ion mass spectrometry) to image individual cells after pulse-chasing with 13 CH
4
and 15 NO
3
, and correlate data on label incorporation with phylogenetic identity of major methane-metabolizing species.