A single centre retrospective analysis of diagnostic fluorescence in
advertisement

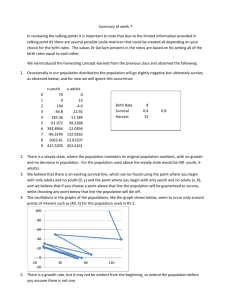
A single centre retrospective analysis of diagnostic fluorescence in situ hybridisation reveals distinct groups of cytogenetic abnormalities, which affect early survival of new myeloma diagnosis. Walker IG 1, Double G 1,2, Milner G2, Kazmi M2, El- Najjar I2, Neat M2,3, Schey S1,4 Streetly M 1,2 1. School of Medicine, King’s College London. 2. Department of Haematology, Guy’s and St Thomas’ Hospital Trust, NHS 3. Department of Cancer Genetics, ViaPath, Guy’s and St Thomas’ Hospital Trust, NHS 4. Department of Haematology, King's College Hospital, London, UK. Background Several specific FISH abnormalities in myeloma have prognostic value with highrisk (t(4;14), t(14;20), 17p-, 1q21) and standard risk (hyperdiploidy) markers identified. However, the prognostic effect of co-associated abnormalities is less well established. We present a multivariate analysis to investigate how cytogenetic abnormalities correlate with each other and whether multiple abnormalities carry a synergistic effect on overall survival (OS). Methods This was a retrospective analysis of newly diagnosed myeloma patients analysed by standard myeloma FISH panel to detect established IgH translocations as well as chromosomal gains and deletions. Detected abnormalities were then analysed by multivariate analysis for correlation and impact on survival. Hierarchical clustering by Ward’s Method allowed abnormalities frequently occurring together to be linked. OS in identified clusters was calculated using the Kaplan-Meier method and Wilcoxon Test was performed to compare OS between groups and patients with ISS stage 3 (ISS3)(i.e. a poor prognosis group). Results 153 newly diagnosed myeloma patients were included in the analysis (median age= 69). Median follow up was 36 months and 3-year survival was 55%. Patients graded ISS3 (n=65) had a median OS of 40 months. Patients with 1q21 copy gains (N=58) had median OS of 41 months. Hyperdiploid patients (n=82) had a 3-year survival of 71%. Cluster analysis identified two groups of abnormalities: Cluster 1 (1q21, Ch13 monosomy, IgH rearrangement with any partner gene) and Cluster 2 (CEP 9,15 Ch5p15.3, CH11q22.3 copy gain) (Figure 1a). 1q21 copy gains alone (n=19) showed a favourable early prognosis when compared to ISS3 or 1q21 gains in association with IgH rearrangement, Ch13 monosomy or both (n=25 & 12 respectively) which was lost with longer follow-up (Figure 1B). 12 month survival of 1q21 gain vs ISS3 was significant at P= 0.026. Within the hyperdiploid cluster (Figure 1C); patients with a greater number of hyperdiploid chromosomes had better OS with a significant difference between ISS 3 patients (p value =0.003). Conclusions This is a small study using Hierarchical cluster analysis that shows: 1. Cytogenetic abnormalities appear to frequently cluster together. 2. 1q21 gain as an isolated abnormality may be associated with better early survival but this trend is lost with longer follow-up. 3. Greater chromosomal hyperdiploidy appears to have a better OS. 4. Further work in a larger patient cohort is required.