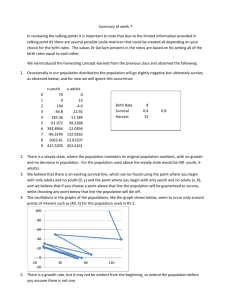
Chapter 4: Analysis of a Single Numerical Variable
advertisement
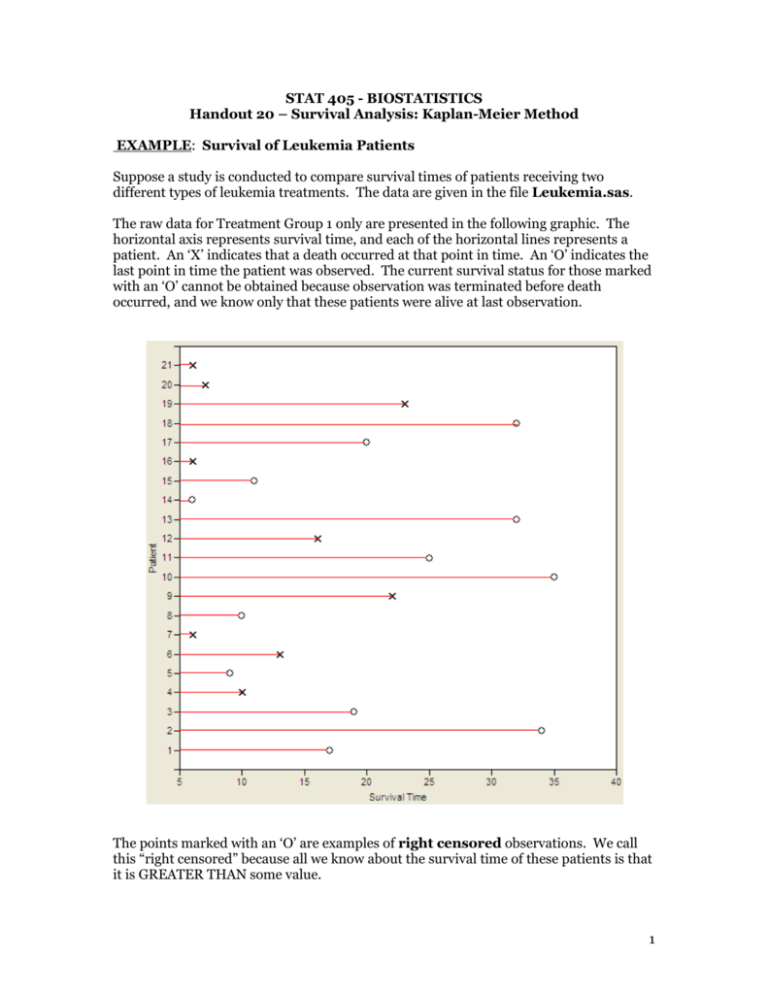
STAT 405 - BIOSTATISTICS Handout 20 – Survival Analysis: Kaplan-Meier Method EXAMPLE: Survival of Leukemia Patients Suppose a study is conducted to compare survival times of patients receiving two different types of leukemia treatments. The data are given in the file Leukemia.sas. The raw data for Treatment Group 1 only are presented in the following graphic. The horizontal axis represents survival time, and each of the horizontal lines represents a patient. An ‘X’ indicates that a death occurred at that point in time. An ‘O’ indicates the last point in time the patient was observed. The current survival status for those marked with an ‘O’ cannot be obtained because observation was terminated before death occurred, and we know only that these patients were alive at last observation. The points marked with an ‘O’ are examples of right censored observations. We call this “right censored” because all we know about the survival time of these patients is that it is GREATER THAN some value. 1 Questions: 1. Using “conventional” statistical methods, how might you approach the analysis of these data? 2. What are the problems with these “conventional” methods? Explain. Survival Analysis Conventional methods discussed earlier in the semester are not appropriate for dealing with either censored data or time-dependent variables. In contrast, survival analysis consists of methods for studying both the occurrence and timing of events, and these methods allow for censoring. Several approaches to survival analysis exist, and we will start by discussing one known as the Kaplan-Meier method. Kaplan-Meier Method In biostatistics, the Kaplan-Meier (KM) estimator is the most widely used method for estimating survivor functions. When there are no censored data, the KM estimator is simple and intuitive. For example, suppose that five patients were observed for six months, and the following was observed: Patient Month in Which Death Occurred 1 1 2 1 3 2 4 4 5 5 2 The survivor function, S(t), is the probability that an event time is greater than t, where t can be any nonnegative number. In this example, an event time refers to the time at which death occurs. Therefore, since there is no censoring, the KM estimator Ŝ(t) is simply the sample proportion of observations which are still alive at each time point: i.e. 𝑆̂(𝑡) = 𝑃(𝑇 ≥ 𝑡) = Time, t # 𝑖𝑛𝑑𝑖𝑣𝑖𝑑𝑢𝑎𝑙𝑠 𝑤𝑖𝑡ℎ 𝑇 ≥𝑡 𝑡𝑜𝑡𝑎𝑙 𝑠𝑎𝑚𝑝𝑙𝑒 𝑠𝑖𝑧𝑒 Ŝ(t) 0 1 2 3 4 5 Kaplan-Meier estimator for censored data When the data are censored and some of the censoring times are smaller than some event times, the observed proportion of cases with event times greater than t can be biased. This happens because cases that are censored before time t may have died before time t, unbeknownst to us. To handle this can consider using the following from conditional probability. Suppose 𝑡𝑘 < 𝑡 ≤ 𝑡𝑘+1 , then 𝑆(𝑡) = 𝑃(𝑇 ≥ 𝑡𝑘+1 ) = 𝑃(𝑇 ≥ 𝑡1 , 𝑇 ≥ 𝑡2 , … , 𝑇 ≥ 𝑡𝑘+1 ) = 𝑃(𝑇 ≥ 𝑡1 ) × ∏𝑘𝑗=1 𝑃(𝑇 ≥ 𝑡𝑘+1 |𝑇 ≥ 𝑡𝑘 ) 𝑘 = ∏[1 − 𝑃(𝑇 = 𝑡𝑗 |𝑇 ≥ 𝑡𝑗 )] 𝑗=1 𝑘 = ∏[1 − 𝜆𝑗 ] where 𝜆𝑗 = 𝑃(𝑑𝑒𝑎𝑡ℎ 𝑎𝑡 𝑡𝑖𝑚𝑒 𝑡𝑗 ) 𝑗=1 3 So 𝑘 𝑆̂(𝑡) ≅ ∏ (1 − 𝑗=1 𝑑𝑗 ) 𝑟𝑗 = ∏ (1 − 𝑗:𝑡𝑗 <𝑡 𝑑𝑗 ) 𝑟𝑗 Here we are assuming there are k distinct event times. At each of these event times tj, there are rj individuals who are said to be “at risk” (they have not yet experienced an event and they have not been censored prior to time tj). Finally, we let dj be the number who experience an event (death) at time tj and let cj denote the number of censored observations between the j-th and (j+1)-st observed event time. Useful identities: 𝑟𝑗 = 𝑟𝑗−1 − 𝑑𝑗−1 − 𝑐𝑗−1 𝑟𝑗 = ∑𝑘≥𝑗(𝑐𝑘 + 𝑑𝑘 ) The product derived above is called the Kaplan-Meier estimator or product-limit estimator and is defined as: 𝑆̂(𝑡) = ∏ (1 − 𝑗:𝑡𝑗 <𝑡 𝑑𝑗 ) 𝑟𝑗 EXAMPLE: Back to Survival of Leukemia Patients For the 21 patients who received Treatment 1, the survival times of the patients are given as follows: 6, 6, 6, 6+, 7, 9+, 10, 10+, 11+, 13, 16, 17+, 19+, 20+, 22, 23, 25+, 32+, 32+, 34+, 35+ We can use the KM method to estimate the survivor function for Treatment Group 1: Time tj # at risk nj # of events dj # censored cj Estimated Probability of Surviving dj 1 n j Estimated Survivor Function Ŝ(t) dj j 1 n j:t j t 4 Using SAS for Kaplan-Meier Estimation of the Survivor Function First, note that the data set should be constructed as follows: data Leukemia; input group duration censor; datalines; 1 6 1 1 6 1 1 6 1 1 6 0 1 7 1 1 9 0 . . 5 For each case in the data set, there must be one variable in the data set which contains either the time that an event occurred or, for censored cases, the last time at which that case was observed. A second variable is necessary if some of the cases are censored. It is common to set this equal to 1 for uncensored cases and 0 for censored cases. Specifically for our data set, the variable duration gives the time in months from the beginning of the study to either death or censoring. The variable censor has a value of 1 for those who died and a value of 0 for those who were censored. An additional variable, group, is an indicator variable for Treatment 1 vs. Treatment 2 patients. To get the KM estimator, you can use PROC LIFETEST as follows: proc lifetest data=leukemia method=km plots=(s); where group=1; time duration*censor(0); symbol1 v=none; run; Comments regarding the ‘time’ statement: The first variable, duration, is the time of event or censoring. The second variable, censor, contains information on whether or not the observation was censored. The number in parentheses is the value of the second variable corresponding to censored observations. Other comments: plots=(s) requests a plot of the survival function vs. time. symbol1 v=none suppresses a symbol ordinarily placed at the data point on the graph. The output is given as follows: 6 Each of the lines in the above output corresponds to one of the 21 cases (except for the first line, which is for time 0). Note that censored cases are marked with an asterisk. The second column gives the KM estimator of the survivor function. When observations share a common event or censoring time, the KM estimate is reported for only the last case; furthermore, no estimates are reported for censored times. Note that although the KM estimator does not appear for all times, it is defined for any time between 0 and the largest event or censoring time. It’s just that it only changes at an observed event time. Questions: 1. What is the estimated probability that a patient will survive for 16 months or more? 2. What is the estimated survival probability for any time from 10 months up to (but not including) 13 months? The survivor function is graphed below: 7 SAS also returns the following: Here, the 25th percentile refers to the smallest event time such that the probability of dying earlier is greater than 25%. No value is reported for the 75th percentile because the KM estimator for these data never reaches a failure probability greater than 55.18% or a survival probability lower than 44.82%. Note that the 50th percentile represents the median death time (here, it is 23 months). SAS also provides an estimated mean time of death, but this estimate is biased downward since there are censoring times greater than the largest event time. Even if this is not the case, when a substantial number of cases are censored, the median is a much preferred measure of central tendency for censored survival data. Definition for Mean, Medians, and Quantiles based on the KM Estimators Mean – ∑𝑘𝑗=1 𝑡𝑗 𝑃(𝑇 = 𝑡𝑗 ) Median – by definition, this is the time 𝜏 such that 𝑆(𝜏) = 0.5. However, in practice, it is defined as the smallest time such that 𝑆(𝜏) ≤ 0.5. The median is more appropriate for censored survival data than the mean. Lower quartile – the smallest time (𝑄1 ) such that 𝑆(𝑄1 ) ≤ 0.75 Upper quartile – the smallest time (𝑄3 ) such that 𝑆(𝑄3 ) ≤ 0.25 Constructing Confidence Intervals for the KM estimators Note that SAS also provided us with a standard error for the “survival” at each point in time. These standard errors can be used to construct confidence intervals, which are easily obtained using the ‘outsurv=’ option in PROC LIFETEST. proc lifetest data=leukemia method=km outsurv=a; where group=1; time duration*censor(0); run; proc print; run; 8 ̂(𝒕)) Estimating the 𝒗𝒂𝒓(𝑺 Uses the delta method which says, if Y ~𝑁(𝜇, 𝜎 2 ) then 𝑔(𝑌)is approximately normally distributed with mean 𝑔(𝜇) and variance [𝑔′ (𝜇)]2 𝜎 2 . For example if we consider 𝑔(𝑌) = log(𝑌) then 1 2 𝑔(𝑌)~𝑁(𝑔(𝜇), [𝑔′ (𝜇)]2 𝜎 2 ) or 𝑔(𝑌)~𝑁(log(𝜇) , (𝜇) 𝜎 2 ) and if 𝑔(𝑌) = 𝑒 𝑌 then 𝑔(𝑌)~𝑁(𝑒 𝜇 , [𝑒 𝜇 ]2 𝜎 2 ) Instead estimating the 𝑣𝑎𝑟(𝑆̂(𝑡)) we can use the delta method to approximate the 𝑣𝑎𝑟(log(𝑆̂(𝑡)). The log(𝑆̂(𝑡))=∑𝑗:𝑡 <𝑡 log(1 − 𝜆̂𝑗 ) and using independence of the 𝜆̂𝑗 ′𝑠 we have 𝑗 𝑣𝑎𝑟[log(𝑆(𝑡))] = ∑ 𝑣𝑎𝑟[log(1 − 𝜆̂𝑗 )] 𝑗:𝑡𝑗 <𝑡 = ∑ ( 𝑗:𝑡𝑗 <𝑡 1 1 − 𝜆̂𝑗 = ∑ ( 𝑗:𝑡𝑗 <𝑡 = ∑ 𝑗:𝑡𝑗 <𝑡 = ∑ 𝑗:𝑡𝑗 <𝑡 2 ) 𝑣𝑎𝑟(𝜆̂𝑗 ) 1 1 − 𝜆̂𝑗 2 ) 𝜆̂𝑗 (1 − 𝜆̂𝑗 )/𝑟𝑗 𝜆̂𝑗 (1 − 𝜆̂𝑗 )𝑟𝑗 𝑑𝑗 (𝑟𝑗 − 𝑑𝑗 )𝑟𝑗 9 Now 𝑆(𝑡) = exp [𝑙𝑜𝑔 (𝑆̂(𝑡))] thus by the delta method again we have 2 𝑣𝑎𝑟 (𝑆̂(𝑡)) = [𝑆̂(𝑡)] 𝑣𝑎𝑟 [log (𝑆̂(𝑡))] thus, 𝑑𝑗 2 𝑣𝑎𝑟 (𝑆̂(𝑡)) = [𝑆̂(𝑡)] ∑ 𝑗:𝑡𝑗 <𝑡 (𝑟𝑗 − 𝑑𝑗 )𝑟𝑗 This is called Greenwood’s Formula. The square root of this variance approximation gives approximate standard errors for the estimated survivor function, i.e. 𝑆𝐸(𝑆̂(𝑡)). Using this standard error we can compute a 95% CI for S(t) as: 𝑆̂(𝑡) ± 1.96 ∙ 𝑆𝐸[𝑆̂(𝑡)] This can yield values outside the range [0,1]. A better approach is to exploit the delta method even further to obtain a CI for 𝐿(𝑡) = log[− log(𝑆(𝑡))]. Since this quantity is unrestricted, the confidence interval will be in the proper range when we transform back. Log-log Approach for Confidence Intervals 1) Define 𝐿(𝑡) = log(− log(𝑆(𝑡))) 2) Form a 95% CI for L(t) 𝐿̂(𝑡) ± 1.96 ∙ 𝑆𝐸(𝐿̂(𝑡)) 3) Since 𝑆(𝑡) = exp(− exp[𝐿(𝑡)]), the 95% CI for S(t) based on the CI for L(t) is given by [exp(−𝑒 (𝐿̂(𝑡)+𝐴) ) , exp(−𝑒 𝐿̂(𝑡)−𝐴 )] 4) Substituting 𝐿̂(𝑡) = log(− log (𝑆̂(𝑡))) into the upper and lower bounds gives confidence limits 𝑒𝐴 𝑒 −𝐴 ([𝑆̂(𝑡)] , [𝑆̂(𝑡)] ) 10 What is 𝑆𝐸(𝐿̂(𝑡))? Using the delta method again… 2 𝑣𝑎𝑟 (𝐿̂(𝑡)) = 𝑣𝑎𝑟[𝑙𝑜𝑔(−log(𝑆̂(𝑡)))] = ( 1 log (𝑆̂(𝑡)) ) ∑ 𝑗:𝑡𝑗 <𝑡 𝑑𝑗 (𝑟𝑗 − 𝑑𝑗 )𝑟𝑗 and the standard error is the square root of this estimated variance. Constructing Confidence Bands for the Survivor Function The pointwise confidence interval for the survivor function S(t) discussed above is valid for ONLY a SINGLE FIXED TIME at which the inference is to be made. In some cases, it is of interest to find the upper and lower confidence bands that guarantee, with a given confidence level, that the survivor function falls within the band for all t in some interval. One such approach was proposed by Hall and Wellner and is implemented in SAS as follows: ods html; ods graphics on; proc lifetest data=leukemia method=km outsurv=a plots=survival(cl cb=hw); where group=1; time duration*censor(0); run; ods graphics off; ods html close; 11 12 Testing for Differences in Survivor Functions Now, let’s consider the observations from both treatment groups 1 and 2. To determine whether the survivor functions differ across treatment, we test the following hypotheses: Ho: S1(t) = S2(t) for all t Ha: S1(t) ≠ S2(t) for all t SAS provides three test statistics for this alternative hypothesis: proc lifetest data=leukemia method=km plots=(s); time duration*censor(0); strata group; symbol1 v=none; symbol2 v=none; run; Note that the likelihood ratio statistic is calculated under the assumption that the observations follow an exponential distribution, which is not always the case! Also, note that SAS PROC LIFETEST returns survivor tables for both treatments (see the table for treatment 2 below; the table for treatment 1 is the same as it was before). 13 We also obtain a plot of both survivor functions: 14 1. 00 0. 75 0. 50 0. 25 0. 00 0 5 10 15 20 25 gr oup=1 gr oup=2 30 35 dur at i on S T RA T A : gr oup=1 Ce n s o r e d We can request Hall and Wellner bands for both functions by adding the ods graphics statements and using the ‘plots’ request as shown on page 8: plots=survival(cl cb=hw); 15 Finally, you can modify the ‘plots’ request using the ‘strata=panel’ option to specify that separate plots for each treatment group be organized into panels instead of overlaying them on the same plot. plots=survival(cl cb=hw strata=panel); 16 Kaplan-Meier Estimation and Testing in R For the 21 patients who received Treatment 1, the survival times of the patients are given as follows: 6, 6, 6, 6+, 7, 9+, 10, 10+, 11+, 13, 16, 17+, 19+, 20+, 22, 23, 25+, 32+, 32+, 34+, 35+ [1,] [2,] [3,] [4,] [5,] [6,] [7,] [8,] [9,] [10,] [11,] [12,] [13,] [14,] [15,] [16,] [17,] [18,] [19,] [20,] [21,] Time Censor 6 1 6 1 6 1 6 0 7 1 9 0 10 1 10 0 11 0 13 1 16 1 17 0 19 0 20 0 22 1 23 1 25 0 32 0 32 0 34 0 35 0 > fit = survfit(Surv(Time,Censor)~1,type="kaplan",conf.type="plain") > summary(fit) Call: survfit(formula = Surv(Time, Censor) ~ 1, type = "kaplan", conf.type = "plain") time n.risk n.event survival std.err lower 95% CI upper 95% CI 6 21 3 0.857 0.0764 0.707 1.000 7 17 1 0.807 0.0869 0.636 0.977 10 15 1 0.753 0.0963 0.564 0.942 13 12 1 0.690 0.1068 0.481 0.900 16 11 1 0.627 0.1141 0.404 0.851 22 7 1 0.538 0.1282 0.286 0.789 23 6 1 0.448 0.1346 0.184 0.712 > plot(fit) > title(main="Kaplan-Meier with Greenwood-Based CI's") 17 > fit = survfit(Surv(Time,Censor)~1,type="kaplan",conf.type="log-log") > summary(fit) Call: survfit(formula = Surv(Time, Censor) ~ 1, type = "kaplan", conf.type = "log-log") time n.risk n.event survival std.err lower 95% CI upper 95% CI 6 21 3 0.857 0.0764 0.620 0.952 7 17 1 0.807 0.0869 0.563 0.923 10 15 1 0.753 0.0963 0.503 0.889 13 12 1 0.690 0.1068 0.432 0.849 16 11 1 0.627 0.1141 0.368 0.805 22 7 1 0.538 0.1282 0.268 0.747 23 6 1 0.448 0.1346 0.188 0.680 > plot(fit) > title(main="Kaplan-Meier with Log-Log CI's") 18