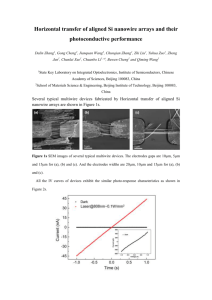
Word - Yan Kou
advertisement

Yan Kou 50 E 98th St, New York, NY 10029 518.572.8255 koukougogo@gmail.com EDUCATION Ph.D. Candidate of Biomedicine, Mount Sinai School of Medicine, Manhattan, NY 2010 - 2014 (expected) Thesis: Computational reconstruction of Autism Spectrum Disorder pathway; Mentor: Avi Ma’ayan, Joseph Buxbaum Author of one book chapter and nine publications in peer-reviewed journals including Nature; Author of four manuscripts in preparation; Invited speaker at two international conferences Awarded Seaver Autism Center Fellowship 2011-2014, $35000/year Certificate of Completion of “Tackling the Challenges of Big Data” course from Massachusetts Institute of Technology M.S. Applied Biotechnology, Uppsala University, Uppsala, Sweden 2008 - 2010 Thesis: Relationship between protein sequence and structure in Frenet representation; Mentor: Antti Niemi B.S. Biotechnology, Beijing Normal University, Beijing, China 2004 - 2008 SKILLS Proficient in applying statistical and non-statistical techniques to large-scale, high-dimensional data; Familiar with supervised and unsupervised machine learning, graph theory and network inference methods Three years experience in programming with Java and Python; Three years experience of performing statistical learning and analysis with MATLAB; Familiar with web development with HTML, CSS and JavaScript; Familiar with Linux shell scripting and cloud-computing; Knowledge of MySQL, PHP and genetic programming Demonstrated communication and presentation skills Fluent in English and Mandarin Chinese, basic level of Swedish EXPERIENCE Mount Sinai School of Medicine, Ph.D. Candidate, Manhattan, New York 2010 - present Research: Developed Support Vector Machine classifiers to predict and prioritize autism susceptible genes among >30,000 human genes with over 2,000 features; Compiled and created a large human protein protein interaction network from 20 databases and developed network-based classifiers using shortest path and mean-first-passage-time algorithm implemented with MATLAB and Python Developed unsupervised clustering method to categorize cancer patients from 103-106 scaled datasets; Implemented interactive web interface for the clustering results using D3.js; Our clustering approach differentiated clinical outcomes better than existing methods and commercial products Constructed formatted data libraries by mining through >2 terabytes of public biological data; Developed a pipeline to process large genomic sequencing data using Java and shell script on computing clusters Teaching: Delivered six Coursera video lectures on Linux, R, cluster-based data processing and genomic sequencing data analysis for Network Analysis in Systems Biology (https://class.coursera.org/netsysbio-001); 25,000+ students enrolled worldwide; lectures viewed 5,000+ times on average and were well received Lectured and served as teaching assistant for Programming for Systems Biology in 2012-2014 General Electric Healthcare, Intern, Uppsala, Sweden 1- 6/2009 Led a student research team and proposed optimization for the novel chromatography product CaptoMMC® Siemens Limited China, Intern and System Administrator, Beijing, China Initiated and designed Point-Of-Sales online channel management system Interpreted real time sales data from SAP system Beijing Pharmaceutical and Biotechnology Center, Intern, Beijing, China Analyzed the trend of global R&D activities of the seed industry 12/2007 - 08/2008 8 -10/2007