Genome-wide association study of sexual maturation in males and
advertisement
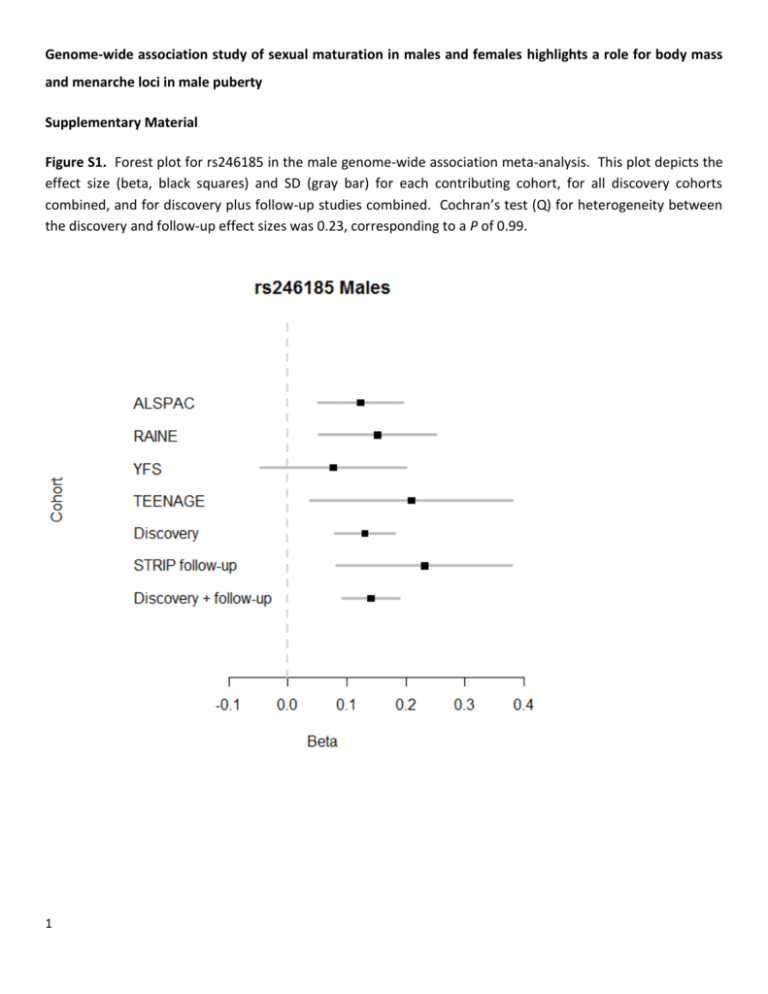
Genome-wide association study of sexual maturation in males and females highlights a role for body mass and menarche loci in male puberty Supplementary Material Figure S1. Forest plot for rs246185 in the male genome-wide association meta-analysis. This plot depicts the effect size (beta, black squares) and SD (gray bar) for each contributing cohort, for all discovery cohorts combined, and for discovery plus follow-up studies combined. Cochran’s test (Q) for heterogeneity between the discovery and follow-up effect sizes was 0.23, corresponding to a P of 0.99. 1 Figure S2. a) QQ plots from the primary meta-analyses. P-values have been automatically double corrected for genomic inflation by the meta-analysis program GWAMA (1, 2) as described in the Materials and Methods. b) Manhattan plots 2 Figure S3. a) Regional plot of the chromosome 16p13.12 locus before follow-up analysis showing the P-value for each marker from the discovery male analysis along the chromosome (genome build 36). rs246185 is highlighted as a yellow triangle and lies within two recombination hotspots between MKL2 and PARN. 3 b) Regional association plot of the same locus imputed against the 1000 Genomes reference set (genome build 37). rs246185 is presented as a yellow triangle, and rs193536 is shown as a red triangle. 4 Figure S4. Position weight matrices (PWMs) representing preferred binding motif sequences for predicted binding sites affected by two SNPs in LD with rs246185 from the RegulomeDB output (3). The red boxes surround rs74755650 in panels a and b, and rs193536 in panels c and d. R2 between rs246185 and rs7475560 is 0.75, and between rs246185 and rs193536 is 0.85. 5 Figure S5. Scatter plots showing the relationship between the effect size for all 31 known BMI-increasing alleles (4) and the effect size for Tanner staging in males, females, and both sexes combined. See Table S6 for the specific alleles that are represented here. 6 Table S1. Cohort summary information (separate excel sheet). Table S2. Genes nearby the chr 16p13.12 locus, their function, and the phenotypic effect of mutants in human and mouse. Approximate distance to rs246185a 32 kb Functionb Mutations in humansc Phenotype in moused blood vessel morphogenesis, cardiac muscle tissue development, cell differentiation, embryonic organ development, heart morphogenesis none cellular, growth/size, mortality/aging, nervous system poly(A)-specific ribonuclease (deadenylation nuclease) bifunctional apoptosis inhibitor 131 kb 3'-exoribonuclease none none 329 kb anti-apoptosis none none ERCC4 excision repair crosscomplementing rodent repair deficiency, complementation group 4 347 kb nucleotide excision repair cellular, growth/size, liver/biliary, mortality/aging PLA2G10 phospholipase A2, group X 369 kb catalyzes the release of fatty acids from phospholipids and play a role in a wide range of physiologic functions. GnRH pathway, MAPK pathway xeroderma pigmentosum complementation group F (XP-F), or xeroderma pigmentosum VI (XP6) none Gene Full name MKL2 myocardin-like 2 PARN BFAR a Estimated from Ensembl (http://www.ensembl.org). Function summarized from AceView (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/index.html?human). c Human mutations queried from Online Mendelian Inheritance in Man (http://www.ncbi.nlm.nih.gov/omim). d Mouse mutations queried from Mouse Phenome Database (http://phenome.jax.org/). b 7 hematopoietic, homeostasis, immune; endocrine/exocrine, homeostasis; cardiovascular, cellular, hematopoietic, homeostasis, immune, muscle, reproductive Table S3. RegulomeDB (1, 2) (http://regulome.stanford.edu/) functional analysis of SNPs in high LD (r2>0.6) with index SNP rs246185 from the 1000 Genomes Pilot I data set. Queried on April 25, 2013. See the website for detailed annotation of analysis, data output, and scoring. SNP Distancea R2 D´ Positionb Regulome DB scorec Evidence rs246185 0 1.000 1.000 14302933 4, Minimal binding evidence rs74755650 356 0.750 0.948 14302577 2b, Likely to affect binding rs181766 554 0.958 1.000 14302379 4, Minimal binding evidence rs193536 588 0.846 1.000 14302345 2b, Likely to affect binding rs246180 3509 0.958 1.000 14299424 No data Chromatin_Structure|FAIRE, Chromatin_Structure|DNase-seq, Protein_Binding|ChIP-seq|EGR1, Protein_Binding|ChIP-seq|SMARCB1, Protein_Binding|ChIP-seq|NR3C1 Motifs|PWM|WT1, Motifs|PWM|MAZR, Motifs|Footprinting|WT1, Motifs|Footprinting|MAZR, Chromatin_Structure|FAIRE, Chromatin_Structure|DNase-seq, Protein_Binding|ChIP-seq|POLR2A Chromatin_Structure|FAIRE, Chromatin_Structure|DNase-seq, Protein_Binding|ChIP-seq|SIN3A, Protein_Binding|ChIP-seq|MXI1, Protein_Binding|ChIP-seq|EP300, Protein_Binding|ChIP-seq|MYC, Protein_Binding|ChIP-seq|MAX, Protein_Binding|ChIP-seq|NFKB1, Protein_Binding|ChIP-seq|RAD21, Protein_Binding|ChIP-seq|USF1, Protein_Binding|ChIP-seq|SMC3, Protein_Binding|ChIP-seq|BHLHE40, Protein_Binding|ChIP-seq|JUND, Protein_Binding|ChIP-seq|RFX5, Protein_Binding|ChIP-seq|PAX5, Protein_Binding|ChIP-seq|GTF2F1, Protein_Binding|ChIP-seq|YY1, Protein_Binding|ChIP-seq|ZNF143, Protein_Binding|ChIP-seq|NR3C1, Protein_Binding|ChIP-seq|POLR2A, Protein_Binding|ChIP-seq|FOSL2, Protein_Binding|ChIP-seq|ZBTB7A, Protein_Binding|ChIP-seq|CTCF, Protein_Binding|ChIP-seq|IRF4, Protein_Binding|ChIP-seq|CDX2, Protein_Binding|ChIP-seq|HDAC2 Motifs|PWM|ER, Motifs|Footprinting|Pax-3, Motifs|Footprinting|ER, Motifs|PWM|Pax-3, Chromatin_Structure|FAIRE, Chromatin_Structure|DNase-seq, Protein_Binding|ChIP-seq|SIN3A, Protein_Binding|ChIP-seq|MXI1, Protein_Binding|ChIP-seq|EP300, Protein_Binding|ChIP-seq|MYC, Protein_Binding|ChIP-seq|MAX, Protein_Binding|ChIP-seq|NFKB1, Protein_Binding|ChIP-seq|RAD21, Protein_Binding|ChIP-seq|USF1, Protein_Binding|ChIP-seq|SMC3, Protein_Binding|ChIP-seq|BHLHE40, Protein_Binding|ChIP-seq|JUND, Protein_Binding|ChIP-seq|RFX5, Protein_Binding|ChIP-seq|PAX5, Protein_Binding|ChIP-seq|GTF2F1, Protein_Binding|ChIP-seq|YY1, Protein_Binding|ChIP-seq|ZNF143, Protein_Binding|ChIP-seq|NR3C1, Protein_Binding|ChIP-seq|POLR2A, Protein_Binding|ChIP-seq|FOSL2, Protein_Binding|ChIP-seq|ZBTB7A, Protein_Binding|ChIP-seq|CTCF, Protein_Binding|ChIP-seq|IRF4, Protein_Binding|ChIP-seq|CDX2, Protein_Binding|ChIP-seq|HDAC2 No data rs1704528 6682 0.881 1.000 14296251 6, Minimal binding evidence 8 Motifs|PWM|c-Ets-1, Motifs|PWM|Elk-1, Motifs|PWM|c-Ets-1(p54), Motifs|PWM|Tel-2, Motifs|PWM|NERF1a, Motifs|PWM|c-Ets-2, Motifs|PWM|Ets, Motifs|PWM|FLI1, Motifs|PWM|Etv1 rs1659127 7127 0.837 0.955 14295806 6, Minimal binding evidence rs30153 10955 0.831 0.912 14291978 6, Minimal binding evidence Motifs|PWM|CAC-bindingprotein, Motifs|PWM|MAZR, Motifs|PWM|Klf7, Motifs|PWM|SP1, Motifs|Footprinting|Klf4, Motifs|PWM|Klf4, Motifs|PWM|SP4, Motifs|PWM|Sp1 Motifs|PWM|Zbtb3 rs30152 12785 0.793 0.910 14290148 No data No data rs246177 14664 0.647 1.000 14288269 4, Minimal binding evidence rs246176 14717 0.647 1.000 14288216 4, Minimal binding evidence Chromatin_Structure|DNase-seq, Protein_Binding|ChIP-seq|POLR2A, Protein_Binding|ChIP-seq|CTCF Chromatin_Structure|DNase-seq, Protein_Binding|ChIP-seq|POLR2A rs246175 15284 0.647 1.000 14287649 3a, Less likely to affect binding rs246173 15596 0.602 1.000 14287337 4, Minimal binding evidence a Distance in bp from index SNP rs246185. Position on human genome build 18. c See http://regulome.stanford.edu/about for a detailed explanation of scoring. 9 b Motifs|PWM|Tcfap2c, Motifs|PWM|Tcfap2a, Chromatin_Structure|FAIRE, Chromatin_Structure|DNase-seq, Protein_Binding|ChIP-seq|GATA1, Protein_Binding|ChIP-seq|TFAP2C, Protein_Binding|ChIP-seq|TFAP2A, Protein_Binding|ChIP-seq|SMARCA4, Protein_Binding|ChIP-seq|IKZF1, Protein_Binding|ChIP-seq|EBF1, Protein_Binding|ChIP-seq|TAL1 Motifs|Footprinting|T, Motifs|Footprinting|AP-4, Chromatin_Structure|FAIRE, Chromatin_Structure|DNase-seq, Protein_Binding|ChIP-seq|GATA1, Protein_Binding|ChIP-seq|HNF4A, Protein_Binding|ChIP-seq|MEF2C, Protein_Binding|ChIP-seq|MYC, Protein_Binding|ChIP-seq|TRIM28, Protein_Binding|ChIP-seq|NANOG, Protein_Binding|ChIP-seq|CEBPB, Protein_Binding|ChIP-seq|ELF1, Protein_Binding|ChIP-seq|SMARCB1, Protein_Binding|ChIP-seq|POLR3A, Protein_Binding|ChIP-seq|HSF1, Protein_Binding|ChIP-seq|SMARCA4, Protein_Binding|ChIP-seq|RFX5, Protein_Binding|ChIP-seq|ETS1, Protein_Binding|ChIP-seq|IKZF1, Protein_Binding|ChIP-seq|TBP, Protein_Binding|ChIP-seq|NR3C1, Protein_Binding|ChIP-seq|FOSL2, Protein_Binding|ChIP-seq|WRNIP1, Protein_Binding|ChIP-seq|ZEB1, Protein_Binding|ChIP-seq|GTF3C2, Protein_Binding|ChIP-seq|GATA3, Protein_Binding|ChIP-seq|RXRA, Protein_Binding|ChIP-seq|HDAC2, Protein_Binding|ChIP-seq|THAP1, Protein_Binding|ChIP-seq|SIN3A, Protein_Binding|ChIP-seq|NRF1, Protein_Binding|ChIP-seq|EP300, Protein_Binding|ChIP-seq|NFKB1, Protein_Binding|ChIP-seq|IRF1, Protein_Binding|ChIP-seq|USF1, Protein_Binding|ChIP-seq|BRCA1, Protein_Binding|ChIP-seq|CCNT2, Protein_Binding|ChIP-seq|PAX5, Protein_Binding|ChIP-seq|GTF2F1, Protein_Binding|ChIP-seq|YY1, Protein_Binding|ChIP-seq|GATA2, Protein_Binding|ChIP-seq|IRF4, Protein_Binding|ChIP-seq|CDX2, Protein_Binding|ChIP-seq|POLR3G, Protein_Binding|ChIP-seq|SRF, Protein_Binding|ChIP-seq|BRF1 Table S4. Power calculations for various allele frequency scenarios. These calculations were performed in R (v 2.15.1). n=10000 Allele freq (allele 1/allele 2) Beta 1x10-5 P 1x10-6 5x10-8 n=6000 Allele freq (allele 1/allele 2) Beta 1x10-5 P 1x10-6 5x10-8 n=4000 Allele freq (allele 1/allele 2) Beta 1x10-5 P 1x10-6 5x10-8 10 0.2/0.8 0.05 0.02 0.01 0 0.2/0.8 0.075 0.26 0.14 0.11 0.2/0.8 0.1 0.77 0.63 0.58 0.5/0.5 0.05 0.08 0.04 0.03 0.2/0.8 0.05 0 0 0 0.2/0.8 0.075 0.05 0.02 0.01 0.2/0.8 0.1 0.31 0.18 0.15 0.5/0.5 0.05 0.02 0.01 0 0.5/0.5 0.075 0.21 0.11 0.09 0.5/0.5 0.1 0.73 0.56 0.52 0.2/0.8 0.05 6.00E-04 0 0 0.2/0.8 0.075 0.01 0.005 0.003 0.2/0.8 0.1 0.1 0.04 0.03 0.5/0.5 0.05 0.5/0.5 0.075 0.06 0.02 0.02 0.5/0.5 0.1 0.33 0.19 0.16 0.004 0.0007 0.0005 0.5/0.5 0.075 0.65 0.49 0.44 0.5/0.5 0.1 0.98 0.96 0.95 Table S5. Association of age at menarche SNPs with Tanner sexual maturation. We extracted known menarche-associated variants from the male, female, and combined Tanner analysis, and report their associations here for the menarche-advancing allele. A higher beta for the Tanner association corresponds to earlier puberty. All SNPs from Elks et al (5) unless otherwise noted. The Bonferroni-corrected significance threshold is 0.001 (corrected for assessing 44 loci). SNP Chr Position Gene β P Male β Male P Female β Female P RXRG Menarcheadvancing allele T rs466639 1 163661506 0.011 0.59 0.052 0.15 -0.009 0.72 rs633715 1 176119203 SEC16B C 0.026 0.11 0.024 0.39 0.027 0.17 rs2947411 rs1172294b,c 2 604168 TMEM18 G 0.036 0.04 -0.014 0.64 0.060 0.005 2 25022704 G 0.006 0.67 -0.021 0.36 0.019 0.25 rs17268785 2 56445587 ADCY3, POMC, RBJ CCDC85A A 0.018 0.31 0.005 0.86 0.025 0.26 rs12472911a 2 141944979 LRP1B T 0.017 0.29 0.013 0.64 0.020 0.33 rs17188434 2 156805022 NR4A2 C 0.028 0.30 0.027 0.58 0.029 0.39 rs12617311 2 199340810 PLCL1 A 0.026 0.07 -0.015 0.55 0.046 0.01 rs7617480 3 49185736 KLHDC8B C -0.001 0.94 -0.007 0.80 0.002 0.94 rs6762477 3 50068213 RBM6 A -0.026 0.06 -0.054 0.02 -0.012 0.46 rs7642134 3 86999572 VGLL3 A 0.020 0.15 0.018 0.43 0.020 0.22 rs6438424 3 119057512 3q13.32 A 0.001 0.97 -0.014 0.56 0.007 0.65 rs2687729 3 129377916 EEFSEC A -0.009 0.57 0.037 0.15 -0.031 0.09 rs6439371 3 134093442 A -0.005 0.72 -0.019 0.43 0.002 0.90 rs3914188a 3 185492742 TMEM108, NPHP3 ECE2 G 0.008 0.59 -0.020 0.45 0.022 0.24 rs2002675 3 187112262 A 0.000 0.99 -0.001 0.97 0.001 0.97 rs13187289 C 0.028 0.11 0.010 0.75 0.038 0.08 a 5 133877076 TRA2B, ETV5 PHF15 a 5 137735214 KDM3B A 0.033 0.05 0.035 0.20 0.031 0.12 rs4840086 6 100315159 G 0.012 0.39 -0.031 0.19 0.033 0.05 rs7759938 6 105485647 PRDM13, MCHR2 LIN28B T 0.080 3.42E-08 0.063 0.012 0.088 5.57E-07 rs1361108 6 126809293 T 0.026 0.05 -0.001 0.95 0.040 0.02 rs1079866 7 41436618 C6orf173, TRMT11 INHBA C 0.054 0.0049 0.048 0.15 0.058 0.01 rs7821178 8 78256392 PXMP3 A 0.009 0.52 0.049 0.04 -0.011 0.53 rs2090409 9 108006909 TMEM38B A 0.061 1.84E-05 0.075 0.002 0.054 0.002 rs10980926 9 113333455 ZNF483 G -0.008 0.55 -0.015 0.51 -0.005 0.79 rs4929923 11 8595776 TRIM66 T 0.018 0.21 0.014 0.55 0.019 0.26 rs900145 11 13250481 ARNTL T 0.046 0.0016 0.049 0.05 0.045 0.01 11 46009151 PHF21A T 0.009 0.69 0.025 0.53 0.002 0.96 rs10899489 11 77773021 GAB2 C -0.006 0.76 0.051 0.11 -0.036 0.12 rs6589964 11 122375893 BSX A 0.015 0.29 0.028 0.26 0.009 0.60 rs9555810a 13 110979438 C 0.034 0.02 0.026 0.31 0.039 0.03 rs6575793 14 100101970 C13orf16, ARHGEF7 BEGAIN T -0.008 0.57 -0.013 0.60 -0.006 0.74 rs3743266a,d 15 58568805 RORA C 0.048 0.0008 0.033 0.18 0.055 0.002 15 65489961 IQCH C -0.004 0.75 -0.033 0.15 0.011 0.52 rs757647 rs16938437 a rs7359257 11 a rs1659127 rs4788196 b 16 14295806 MKL2 G 0.050 0.0008 0.117 4.35E-06 0.016 0.37 16 29874935 MAPK3 G 0.016 0.24 0.016 0.49 0.016 0.33 rs9939609 16 52378028 FTO A 0.028 0.04 0.003 0.91 0.040 0.01 rs1364063 16 68146073 NFAT5 T 0.014 0.30 0.022 0.35 0.010 0.54 rs9635759 17 46968784 CA10 G 0.017 0.25 -0.039 0.13 0.046 0.01 rs2243803 18 41210670 SLC14A2 T 0.023 0.09 0.017 0.48 0.026 0.12 rs1398217 18 43006236 FUSSEL18 G 0.001 0.93 -0.011 0.65 0.007 0.67 rs1862471 19 9861322 OLFM2 G 0.023 0.10 0.030 0.22 0.020 0.25 rs10423674 19 18678903 CRTC1 C 0.031 0.03 0.002 0.95 0.045 0.01 rs852069 20 17070593 PCSK2 A 0.006 0.66 -0.011 0.63 0.015 0.37 a a a Defined as a possible menarche locus (5). Associated with menarche in a study of the pubertal growth spurt (6). c Associated with menarche in a study of the overlap between body mass loci and menarche (7). d Associated with menarche in African Americans (8). b 12 Table S6. Association of body mass index-increasing alleles to Tanner sexual maturation. We extracted known BMI-associated variants from the Tanner male, female, and combined analyses, and report their associations here for the BMI-increasing allele. A higher Tanner beta corresponds to earlier pubertal timing. All SNPs are from (4) unless otherwise noted. The Bonferroni-corrected significance threshold is 0.0016 (corrected for assessing 31 loci). SNP Chr Position Gene BMIincreasing allele β P Male β Male P Female β Female P BMI loci previously associated with age at menarchea rs2815752 1 72585028 NEGR1 A -0.001 0.93 -0.026 0.27 0.011 0.52 1 74764232 TNNI3K A 0.024 0.07 0.055 0.02 0.009 0.60 1 176180142 SEC16B T -0.025 0.12 -0.030 0.29 -0.023 0.25 b 2 612827 TMEM18 C 0.037 0.04 -0.016 0.6 0.062 0.004 b,c 2 25011512 C 0.006 0.67 -0.023 0.31 0.020 0.22 rs887912 2 59156381 ADCY3/ POMC/RBJ FANCL T -0.004 0.78 -0.070 0.005 0.031 0.09 rs9816226 3 187317193 ETV5 T 0.012 0.51 -0.028 0.36 0.032 0.15 rs10938397 4 44877284 GNPDA2 G 0.017 0.2 0.005 0.82 0.023 0.16 rs987237 6 50911009 TFAP2B G -0.005 0.77 0.004 0.88 -0.010 0.64 rs4929949 11 8561169 RPL27A/STK33 C 0.033 0.02 0.039 0.09 0.029 0.07 rs10767664 11 27682562 BDNF A 0.023 0.17 0.039 0.17 0.015 0.47 rs7138803 12 48533735 FAIM2 A 0.028 0.04 0.037 0.11 0.023 0.16 rs12444979 16 19841101 GPRC5B C 0.058 0.003 0.085 0.01 0.044 0.06 b 16 52361075 FTO A 0.024 0.08 -0.011 0.62 0.041 0.01 19 39001372 KCTD15 G 0.004 0.75 0.032 0.19 -0.009 0.6 rs1514175 b rs10913469b rs2867125 rs713586 a rs1558902 rs29941 BMI loci not associated with age at menarche rs1555543 1 96717385 PTBP2 C -0.011 0.43 -0.016 0.5 -0.008 0.62 rs2890652 2 142676401 LRP1B C 0.027 0.12 0.010 0.72 0.035 0.1 rs13078807 3 85966840 CADM2 G 0.001 0.97 -0.026 0.36 0.014 0.49 rs2112347 5 75050998 FLJ35779 T 0.015 0.29 0.040 0.09 0.002 0.92 rs206936 6 34410847 NUDT3 G 0.016 0.33 0.010 0.73 0.019 0.34 9 28404339 LRRN6C G 0.002 0.91 0.023 0.36 -0.009 0.61 rs3817334 11 47607569 MTCH2 T 0.042 0.002 0.015 0.51 0.055 0.0008 rs4771122 13 26918180 MTIF3 G 0.005 0.74 0.025 0.37 -0.004 0.84 rs9568856d 13 52962982 OLFM2 A 0.016 0.44 0.005 0.89 0.021 0.4 rs11847697 14 29584863 PRKD1 T 0.035 0.30 0.041 0.5 0.032 0.43 rs10150332 14 79006717 NRXN3 C -0.004 0.79 0.040 0.15 -0.026 0.18 rs2241423 15 65873892 MAP2K5 G 0.006 0.72 0.029 0.28 -0.006 0.75 rs7359397 16 28793160 SH2B1 T 0.011 0.41 0.014 0.54 0.01 0.55 b 18 55990749 MC4R A -0.042 0.008 -0.090 0.0009 -0.018 0.36 rs2287019 19 50894012 QPCTL C -0.03 0.09 -0.006 0.84 -0.042 0.05 rs3810291 19 52260843 TMEM160 A 0.011 0.48 -0.010 0.71 0.022 0.26 rs10968576 c rs571312 a Associated with age at menarche in (5) and/or (7). Also associated with or in LD (r2>0.9) with a SNP associated with childhood BMI (9). c Also associated with decreased pubertal growth across puberty (relative pubertal height change from 8 to adult) (6). d Childhood BMI locus (9). b 13 Table S7. MAGENTA gene set enrichment analyses. See Materials and Methods and Segrè et al (10) for a detailed description of the method. 95th percentile males and females combined Database Gene set Centromere DNA-binding protein 10 Nominal GSEA Pvalue 9.00 E-04 ligand-dependent nuclear receptor activity KEGG ACUTE MYELOID LEUKEMIA positive regulation of transcription from RNA polymerase II promoter 27 1.00E-04 0.23 1 7 NR4A2, RORA 52 265 6.00E-04 7.00E-04 0.12 0.83 3 13 9 26 MAPK3 ARNTL, INHBA, NR4A2, PROP1, RORA, RXRG, VEGFA, CRTC1, MKL2, BSX embryonic pattern specification post-Golgi vesicle-mediated transport steroid hormone receptor activity nuclear hormone receptor 9 38 8.00E-04 2.80E-03 0.18 0.97 0 2 4 7 44 44 4.90E-03 5.60E-03 0.97 0.28 2 2 7 7 phosphoprotein phosphatase activity Signal transduction 33 248 5.70E-03 5.80E-03 0.96 1.00 2 12 6 22 Transmission across chemical synapses Panther Oxytocin receptor mediated signaling pathway REACTOME Nuclear receptor transcription pathway GOTERM dephosphorylation GOTERM 3',5'-cyclic-AMP phosphodiesterase activity th 75 percentile males and females combined Database Gene set 121 6.00E-03 0.46 6 13 NR4A2, RORA, RXRG, PLA2G10, TRIB1 ADCY3 16 7.40E-03 0.34 1 4 PLCL1, PLCG1 47 8.30E-03 0.71 2 7 NR4A2, RORA, RXRG 17 9 8.40E-03 8.70E-03 1.00 1.00 1 0 4 3 Gene set size Nominal GSEA Pvalue FDR Expected # genes Observed # genes GOTERM aminopeptidase activity 25 4.00E-06 0.004 6 17 REACTOME Hormone sensitive lipase HSL mediated triacylglycerol hydrolysis KEGG APOPTOSIS 12 4.20E-05 0.004 3 10 74 1.00E-04 0.01 19 34 PANTHER MOLECULAR FUNCTION GOTERM KEGG GOTERM GOTERM GOTERM GOTERM PANTHER MOLECULAR FUNCTION GOTERM PANTHER BIOLOGICAL PROCESS REACTOME KEGG 14 Gene set size FDR Expected # genes Observed # genes 0.02 1 4 Flagged gene names NR4A2, RORA, RXRG NR4A2, RORA, RXRG Flagged gene names GOTERM manganese ion binding 25 1.00E-04 0.11 6 15 REACTOME INTRINSIC PATHWAY FOR APOPTOSIS 28 1.30E-03 0.13 7 15 BIOCARTA AGPCR PATHWAY 13 1.40E-03 0.10 3 9 BIOCARTA CSK PATHWAY 19 1.90E-03 0.12 5 11 REACTOME APOPTOSIS 113 2.20E-03 0.24 28 42 GOTERM steroid hormone receptor activity 44 2.40E-03 0.84 11 20 NR4A2, RORA, RXRG REACTOME 47 2.70E-03 0.25 12 21 NR4A2, RORA, RXRG 53 2.90E-03 0.17 13 23 CETP GOTERM NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY LPS IL-1 Mediated Inhibition of RXR Function apoptosis 419 3.00E-03 0.73 105 129 ARHGEF7, LY86, NISCH, FAIM2, BFAR GOTERM cation binding 28 3.10E-03 0.65 7 14 GOTERM transcription factor activity 786 3.30E-03 0.64 197 228 GOTERM autophagic vacuole 10 3.30E-03 0.76 3 7 GOTERM actin binding 250 3.50E-03 0.60 63 81 GOTERM internal side of plasma membrane 13 4.00E-03 0.74 3 8 BIOCARTA P53 HYPOXIA PATHWAY 20 4.00E-03 0.16 5 11 PANTHER_MOLECULA R_FUNCTION GOTERM Epimerase/racemase 35 4.60E-03 0.77 9 16 13 5.60E-03 0.63 3 8 BIOCARTA positive regulation of cytokine secretion MYOSIN PATHWAY 29 5.90E-03 0.21 7 14 ARHGEF7 KEGG KEGG PROSTATE CANCER 84 6.10E-03 0.41 21 32 FGFR1, MAPK3 REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE 1 cell development 59 6.50E-03 0.40 15 24 ITPR2 16 7.60E-03 0.59 4 9 ligand-dependent nuclear receptor activity Detoxification 27 7.80E-03 0.64 7 13 57 7.80E-03 1.00 14 23 TELOMERE MAINTENANCE 45 8.80E-03 0.42 11 19 microtubule bundle formation 9 9.00E-03 0.75 2 6 Ingenuity GOTERM GOTERM PANTHER BIOLOGICAL PROCESS REACTOME GOTERM 15 ARNTL, KLF9, ETV5, HOXC13, MAF, NR4A2, PAX5, PROP1, RORA, RXRG, TBX15, TFAP2B, LHX3, HESX1, NFE2L3, NFAT5, ZNF483, BSX KCNMA1, MKL2 NR4A2, RORA GOTERM peroxisomal membrane 39 9.10E-03 0.67 10 17 GOTERM telomere maintenance via telomerase Insulin/IGF pathway- mitogen activated protein kinase kinase/MAP kinase cascade 9 9.60E-03 0.66 2 6 19 9.70E-03 0.60 5 10 Gene set size FDR Expected # genes Observed # genes 0.05 1 4 Panther 95th percentile females Database Gene set Thyrotropin-releasing hormone receptor signaling pathway regulation of transcription, DNAdependent 11 Nominal GSEA Pvalue 1.40E-03 816 1.00E-04 0.43 41 64 9 5.00E-04 0.14 0 4 GOTERM histone methyltransferase activity (H3-K4 specific) protein kinase cascade 85 6.00E-04 0.42 4 13 GOTERM zinc ion binding 1488 1.30E-03 0.55 74 96 GOTERM dendrite development 19 1.70E-03 0.62 1 5 KLF9, NR4A2, RORA, RXRG, SF1, LHX3, BRAP, ZNF259, RBM6, PHF15, BFAR, PHF21A, FANCL, ADAMTS9, ZNF608, PRDM13, ZNF483, LIN28B BDNF GOTERM inhibition of adenylate cyclase activity by G-protein signaling pathway Transcription_cofactor 29 2.10E-03 0.41 1 6 ADCY3 136 3.00E-03 0.43 7 15 SF1 associative learning 13 3.00E-03 0.37 1 4 REACTOME G_ALPHA_Z_SIGNALLING_EVENTS 14 3.10E-03 0.43 1 4 GOTERM microtubule associated complex 24 4.90E-03 0.48 1 5 GOTERM transcription factor activity 789 5.20E-03 0.59 39 55 Panther GOTERM GOTERM PANTHER_MOLECULA R_FUNCTION GOTERM 16 Flagged gene names ARNTL, ETV5, HOXC13, MAF, NR4A2, PAX5, PROP1, RORA, RXRG, TBX15, TFAP2B, LHX3, HESX1, NFE2L3, NFAT5, ZNF483, LIN28B, BSX ADCY3 ARNTL, KLF9, ETV5, HOXC13, MAF, NR4A2, PAX5, PROP1, RORA, RXRG, TBX15, TFAP2B, LHX3, HESX1, NFE2L3, NFAT5, ZNF483, BSX GOTERM phosphoprotein phosphatase activity 33 5.30E-03 0.43 2 6 GOTERM histone H3-K4 methylation 8 5.40E-03 0.41 0 3 PANTHER_BIOLOGICAL _PROCESS GOTERM Extracellular_transport_and_import 66 5.80E-03 0.90 3 9 4 iron, 4 sulfur cluster binding 25 6.50E-03 0.47 1 5 CDKAL1 REACTOME PKA_ACTIVATION 17 8.00E-03 0.70 1 4 ADCY3 Panther Circadian_clock_system 9 8.50E-03 0.12 0 3 ARNTL GOTERM activation of protein kinase A activity 17 9.20E-03 0.49 1 4 ADCY3 GOTERM cognition 9 9.40E-03 0.44 0 3 CHD7 Gene set size FDR Expected # genes Observed # genes Flagged gene names ADCY3 th 75 percentile females Database Gene set KEGG KEGG_DILATED_CARDIOMYOPATHY 81 Nominal GSEA Pvalue 1.00E-04 0.01 20 36 KEGG KEGG_APOPTOSIS 75 1.00E-04 0.02 19 33 KEGG KEGG_LONG_TERM_POTENTIATION 64 1.00E-04 0.01 16 29 Panther PI3_kinase_pathway 13 2.00E-04 0.00 3 10 KEGG KEGG_ACUTE_MYELOID_LEUKEMIA 52 8.00E-04 0.02 13 24 MAPK3 KEGG 191 9.00E-04 0.03 48 67 KEGG KEGG_REGULATION_OF_ACTIN_CYT OSKELETON KEGG_ENDOMETRIAL_CANCER 50 9.00E-04 0.02 13 23 FGF8, FGFR1, MAPK3, ARHGEF7 MAPK3 KEGG KEGG_PROSTATE_CANCER 84 2.40E-03 0.05 21 33 FGFR1, MAPK3 GOTERM ligand-dependent nuclear receptor activity TEL_PATHWAY 27 6.00E-04 0.60 7 15 NR4A2, RORA 18 7.00E-04 0.07 5 11 90 9.00E-04 0.23 23 37 GRB14, PLCG1 59 1.10E-03 0.16 15 26 ITPR2 BIOCARTA CELL_SURFACE_INTERACTIONS_AT_T HE_VASCULAR_WALL REGULATION_OF_INSULIN_SECRETIO N_BY_GLUCAGON_LIKE_PEPTIDE_1 BAD_PATHWAY 25 1.20E-03 0.12 6 14 MAPK3 GOTERM phosphoprotein phosphatase activity 33 1.30E-03 0.54 8 17 BIOCARTA CSK_PATHWAY 19 1.80E-03 0.08 5 11 GOTERM histone H2A acetylation 12 1.90E-03 0.52 3 8 REACTOME TELOMERE_MAINTENANCE 47 2.50E-03 0.27 12 21 BIOCARTA CK1_PATHWAY 17 3.10E-03 0.08 4 10 BIOCARTA REACTOME REACTOME 17 ITPR2, MAPK3 GOTERM autophagic vacuole 10 3.10E-03 0.52 3 7 KEGG 73 3.20E-03 0.06 18 29 MAPK3, NFAT5 GOTERM KEGG_B_CELL_RECEPTOR_SIGNALIN G_PATHWAY central nervous system development 92 3.20E-03 0.41 23 35 PROP1, FAIM2, CHD7 GOTERM hormone biosynthetic process 10 3.70E-03 0.45 3 7 REACTOME INHIBITION_OF_INSULIN_SECRETION _BY_ADRENALINE_NORADRENALINE autophagic vacuole membrane 28 3.70E-03 0.26 7 14 12 3.80E-03 0.40 3 8 KEGG_CHEMOKINE_SIGNALING_PAT HWAY response to UV 157 3.90E-03 0.09 39 54 ADCY3, MAPK3 28 3.90E-03 0.54 7 14 ERCC4 Neurotransmitter_release 87 4.00E-03 0.77 22 33 ITPR2, KCNMA1 61 4.10E-03 0.39 15 25 ARHGEF7 121 4.50E-03 0.26 30 43 ADCY3 REACTOME Rho guanyl-nucleotide exchange factor activity TRANSMISSION_ACROSS_CHEMICAL _SYNAPSES DARPP32_EVENTS 26 4.60E-03 0.17 7 13 GOTERM cation binding 29 4.60E-03 0.52 7 14 GOTERM visual learning 23 4.90E-03 0.56 6 12 REACTOME TIE2_SIGNALING 18 5.20E-03 0.24 5 10 GRB14 KEGG KEGG_CALCIUM_SIGNALING_PATHW AY Signal_transduction 158 5.40E-03 0.09 40 54 ADCY3, ITPR2, PLCG1, TACR3 247 5.40E-03 0.47 62 79 NR4A2, RORA, RXRG, PLA2G10, TRIB1 76 5.70E-03 0.10 19 29 18 5.70E-03 0.48 5 10 REACTOME KEGG_HYPERTROPHIC_CARDIOMYO PATHY_HCM hydrogen ion transmembrane transporter activity EXTENSION_OF_TELOMERES 26 6.20E-03 0.19 7 13 BIOCARTA CARM1_PATHWAY 13 6.20E-03 0.11 3 8 GOTERM nuclear body 16 6.40E-03 0.35 4 9 GOTERM microtubule associated complex 24 6.50E-03 0.38 6 12 GOTERM cell migration 54 6.50E-03 0.45 14 22 GOTERM hyaluronic acid binding 16 6.60E-03 0.43 4 9 STAB1 REACTOME DOWNSTREAM_EVENTS_IN_GPCR_SI GNALING 391 6.60E-03 0.22 98 119 ADCY3, GIPR, GNRHR, ITPR2, KISS1, MC4R, POMC, TAC3, TACR3, ARHGEF7, PROK2, GOTERM KEGG GOTERM PANTHER_BIOLOGICAL _PROCESS GOTERM REACTOME PANTHER_BIOLOGICAL _PROCESS KEGG GOTERM 18 PROKR2 GOTERM 79 6.80E-03 0.37 20 30 ARHGEF7 KEGG induction of apoptosis by extracellular signals KEGG_RENAL_CELL_CARCINOMA 67 6.90E-03 0.10 17 26 MAPK3, VEGFA GOTERM growth cone 51 7.30E-03 0.47 13 21 ARHGEF7 GOTERM ER-Golgi intermediate compartment 36 7.80E-03 0.44 9 16 REACTOME DEPOLARIZATION_OF_THE_PRESYNA PTIC_TERMINAL_TRIGGERS_THE_OP ENING_OF_CALCIUM_CHANNELS arachidonic acid secretion 11 7.80E-03 0.21 3 7 11 8.20E-03 0.52 3 7 PANTHER_BIOLOGICAL _PROCESS GOTERM Cell_structure_and_motility 169 8.30E-03 0.38 42 56 ER overload response 9 9.00E-03 0.49 2 6 PANTHER_BIOLOGICAL _PROCESS REACTOME Amino_acid_activation 33 9.20E-03 0.53 8 15 WARS2 45 9.40E-03 0.27 11 19 SLC6A14 GOTERM AMINO_ACID_AND_OLIGOPEPTIDE_ SLC_TRANSPORTERS embryonic pattern specification 9 9.50E-03 0.37 2 6 KEGG KEGG_MELANOMA 65 9.50E-03 0.13 16 25 GOTERM clathrin coated vesicle membrane 11 9.70E-03 0.46 3 7 GOTERM NuA4 histone acetyltransferase complex 14 9.90E-03 0.41 4 8 Gene set Gene set size FDR Expected # genes Observed # genes PANTHER_MOLECULA R_FUNCTION GOTERM Protease_inhibitor 10 Nominal GSEA Pvalue 1.10E-03 0.02 1 4 cytosol 1118 1.00E-04 0.70 56 78 PANTHER_BIOLOGICAL _PROCESS KEGG Apoptosis 173 1.00E-04 0.17 9 20 51 3.00E-04 0.10 3 9 GOTERM KEGG_AMYOTROPHIC_LATERAL_SCL EROSIS_ALS cysteine-type endopeptidase activity 48 1.40E-03 0.80 2 8 REACTOME ERK_MAPK_TARGETS 19 1.60E-03 0.29 1 5 GOTERM 95th percentile males Database 19 FGF8, FGFR1, MAPK3 Flagged gene names GAD2, PLCG1, PRKD1, MAPK3, RPL27A, ARHGEF7, SPRY2, NISCH PRKD1 MAPK3 KEGG KEGG_STEROID_HORMONE_BIOSYN THESIS Other_phosphatase 36 2.10E-03 0.08 2 7 68 2.20E-03 0.19 3 10 28 2.50E-03 0.09 1 6 72 2.50E-03 0.33 4 10 21 2.70E-03 0.28 1 5 REACTOME Amyotrophic.Lateral.Sclerosis.Signali ng P75_NTR_RECEPTOR_MEDIATED_SIG NALLING ABORTIVE_ELONGATION_OF_HIV1_T RANSCRIPT_IN_THE_ABSENCE_OF_T AT SIGNALLING_BY_NGF 202 2.80E-03 0.41 10 20 BIOCARTA CASPASE_PATHWAY 21 3.50E-03 0.39 1 5 GOTERM T cell differentiation in the thymus 13 3.70E-03 1.00 1 4 GOTERM bone resorption 13 3.80E-03 0.81 1 4 REACTOME APOPTOSIS 116 4.30E-03 0.25 6 13 REACTOME NUCLEAR_EVENTS_KINASE_AND_TR ANSCRIPTION_FACTOR_ACTIVATION spliceosomal snRNP assembly 22 4.60E-03 0.26 1 5 24 5.60E-03 0.78 1 5 25 6.00E-03 0.17 1 5 GOTERM Alzheimer_diseaseamyloid_secretase_pathway ubiquitin-specific protease activity 24 6.10E-03 0.86 1 5 GOTERM early endosome membrane 35 7.50E-03 0.76 2 6 BIOCARTA P53_PATHWAY 16 7.60E-03 0.30 1 4 GOTERM cytoskeleton 612 7.70E-03 0.71 31 44 MAPK3, DNM3 GOTERM microtubule 210 7.70E-03 0.72 11 19 SPRY2, DNM3 GOTERM 9 8.00E-03 0.87 0 3 GOTERM regulation of mitochondrial membrane permeability cell-cell adherens junction 25 8.10E-03 0.77 1 5 GOTERM microtubule bundle formation 9 8.80E-03 0.89 0 3 75 percentile males Database Gene set Gene set size FDR Expected # genes Observed # genes GOTERM coreceptor activity 15 Nominal GSEA Pvalue 6.00E-04 0.53 4 10 REACTOME NUCLEOTIDE_LIKE_PURINERGIC_REC EPTORS 11 1.30E-03 0.10 3 8 PANTHER_MOLECULA R_FUNCTION Ingenuity REACTOME REACTOME GOTERM Panther NUDT3 ARHGEF7 ADCY3, IRS1, ITPR2, PLCG1, MAPK3, MAP2K5, ARHGEF7 MAPK3 th 20 Flagged gene names REACTOME P2Y_RECEPTORS 7 1.90E-03 0.15 2 6 Ingenuity T.Cell.Receptor.Signaling 33 2.80E-03 0.14 8 16 Ingenuity 28 3.80E-03 0.09 7 14 GOTERM Amyotrophic.Lateral.Sclerosis.Signali ng positive regulation of transcription 123 4.20E-03 1.00 31 44 MKL2 GOTERM cytosol 1118 4.50E-03 1.00 280 311 GAD2, PLCG1, PRKD1, MAPK3, RPL27A, ARHGEF7, SPRY2, NISCH BIOCARTA AGPCR_PATHWAY 13 4.50E-03 0.58 3 8 REACTOME HIV_INFECTION 167 4.60E-03 0.39 42 57 KEGG KEGG_ARRHYTHMOGENIC_RIGHT_V ENTRICULAR_CARDIOMYOPATHY_AR VC RHO_GTPASE_CYCLE 68 5.10E-03 0.72 17 27 108 5.70E-03 0.38 27 39 ARHGEF7 PANTHER_BIOLOGICAL _PROCESS BIOCARTA Signal_transduction 244 5.80E-03 0.72 61 78 NR4A2, RORA, RXRG, PLA2G10, TRIB1 MCM_PATHWAY 18 6.00E-03 0.32 5 10 GOTERM filopodium assembly 13 6.10E-03 1.00 3 8 REACTOME CD28_DEPENDENT_VAV1_PATHWAY 11 6.20E-03 0.39 3 7 PANTHER_MOLECULA R_FUNCTION GOTERM Annexin 63 6.80E-03 1.00 16 25 nucleotide binding 1614 7.00E-03 1.00 404 434 REACTOME 187 7.20E-03 0.34 47 62 GOTERM REGULATION_OF_INSULIN_SECRETIO N cysteine-type endopeptidase activity 48 7.30E-03 1.00 12 20 BIOCARTA ACH_PATHWAY 16 7.40E-03 0.29 4 9 PANTHER_BIOLOGICAL _PROCESS Other_metabolism 379 9.20E-03 0.70 95 114 REACTOME 21 PLCG1 DNM3 ADCY3, FGFR1, PARN, PRKD1, MAPK3, MAP2K5, SFRS10, WARS2, DNM3, TNNI3K, CHD7, PTBP2, EEFSEC, CPEB4 ITPR2 MAP2K5, MRPS22 Cohort study descriptions Avon Longitudinal Study of Parents and Children (ALSPAC) ALSPAC is a prospective birth cohort which recruited pregnant women with expected delivery dates between April 1991 and December 1992 from Bristol, United Kingdom. 14,541 pregnant women were initially enrolled with 14,062 children born. Detailed information on health and development of children and their parents were collected from regular clinic visits and completion of questionnaires. A detailed description of the cohort has been published previously (11). Ethical approval was obtained from the ALSPAC Law and Ethics Committee and the Local Ethics Committees. Please note that the study website contains details of all the data that is available through a fully searchable data dictionary (http://www.bris.ac.uk/alspac/researchers/dataaccess/data-dictionary/). A total of 9,912 subjects were genotyped using the Illumina HumanHap550 quad genome-wide SNP genotyping platform by 23andMe subcontracting the Wellcome Trust Sanger Institute, Cambridge, UK and the Laboratory Corporation of America, Burlington, NC, USA. An important facet in the study of children as they go through late childhood into adolescence concerns the timing of the onset of puberty. One of the measures used to assess the stage of puberty in the ALSPAC study is concerned with a self-rating of puberty. The questionnaire was developed in association with Dr Carol Rubin of the Centers for Disease Control (CDC), Atlanta, USA. CDC funded the printing and coding of these questionnaires. The questions asked were obviously different for boys and girls. Each questionnaire included a set of pictures, based on those developed by Tanner adapted from those used in studies in the USA. Individuals were excluded from further analysis on the basis of having incorrect gender assignments; extreme heterozygosity (<0.320 and >0.345 for the Sanger data and <0.310 and >0.330 for the LabCorp data); high levels of individual missingness (>3%); evidence of cryptic relatedness (>10% IBD) and being of non-European ancestry (as detected by a multidimensional scaling analysis seeded with HapMap 2 individuals). EIGENSTRAT analysis revealed no additional obvious population stratification and genome-wide analyses with other phenotypes indicate a low lambda. The resulting data set consisted of 8,365 individuals. SNPs with a minor allele frequency of <1% and call rate of <95% were removed. Only SNPs which passed an exact test of Hardy– Weinberg equilibrium (P >5 × 10-7) were considered for analysis. After cleaning, 500,527 SNPs were available for analysis. Known autosomal variants were imputed with MACH 1.0.16 Markov Chain Haplotyping software (12), using CEPH individuals from phase 2 of the HapMap project (HG18) as a reference set (release 22). For the X chromosomal variants, imputation was performed using MiniMac (v4.43) (13) and CEPH individuals from phase 3 of the HapMap project (HG18) were used as the reference set. 1958 British Birth Cohort or NCDS (B58C-WTCCC) and 1958 British Birth Cohort or NCDS (B58C-T1DGC): The population-based 1958BC included initially all children born in England, Scotland or Wales during one week in March 1958 (N=17,415). Medical examination was carried out at age 11 years (1969), and this included an assessment of pubertal development based on breast rating in participating girls. DNA was collected as part of a biomedical examination carried out at 45 years. Genome-wide data for the 1958BC was obtained through two sub-studies in which participants of the 1958BC members (restricted to white European ancestry) were used as a control population. First, 3,000 cohort members were randomly selected as controls for the 22 Wellcome Trust Case Control Consortium (WTCCC (14) including 1,083 girls with Tanner staging) and genotyped on the Affymetrix SNP 6.0 platform. Secondly, 2,592 participants (1083 girls with staging) were used as controls for a type 1 diabetes case-control study (T1DGC (15)), with samples genotyped through the JDRF/WT Diabetes and Inflammation Laboratory (DIL) using the Illumina Infinium 550K chip. Ethical approval for the biomedical survey was obtained from the South East Multi-centre Research Ethics Committee (ref. 01/1/44) and the Joint UCL/UCLH Committees on the Ethnics of Human Research (Committee A) (ref. 08/H0714/40). Cardiovascular Risk in Young Finns Study (YFS): The Cardiovascular Risk in Young Finns (YFS) is a populationbased 30 year follow up-study (http://youngfinnsstudy.utu.fi/). The first cross-sectional survey was conducted in 1980, when 3,596 Caucasian subjects aged 3-18 years participated. The follow-up studies for the same subjects were performed in 1983, 1986, and 2007. In adulthood, the latest 30-year follow-up study was conducted in 2011 (ages 33-48 years) with ca. 2,100 participants. The study cohort for the present analysis comprised of subjects who had data on Tanner pubertal stage classification at 9 to 18 years old (measured in 1980, 1983 and 1986) available with genotype and other risk factor data (16). The study was approved by the local Ethical Committees and was performed according to the Helsinki Declaration. Netherlands Twin Register (NTR): The Netherlands Twin Register (NTR) is a large population-based study that has collected data on the health and behaviour of twin families since 1986 (17). Data on Tanner stages have been collected in a number of subprojects. In some, Tanner stage was assessed by a clinician or trained researcher; in others, Tanner stage was based on self-reports using pictures or schematic drawings (18, 19). Blood and/or buccal samples were collected in several projects and genotyped on the Affymetrix Human SNP Array 6.0 at the Avera Institute, Sioux Falls, South Dakota (USA) (20). Genotypes were called using the BIRDSEED V2 algorithm and SNPs were imputed at the MD Anderson Cancer Center, Houston, Texas (USA) using the BEAGLE software. If an individual had multiple samples available, the sample with the highest quality was selected. In monozygotic twin pairs, one individual was selected at random. Siblings and dizygotic twin pairs were kept in the dataset; in total, genotypic and phenotypic data were available for 265 children. To correct the analyses for the dependencies in the data, the --within option in Plink was used with family number as a cluster variable. Study protocols were approved by the medical ethics board of the VU Medical Center Amsterdam, the Netherlands (IRB number IRB00002991). Netherlands Study of Depression and Anxiety (NESDA) and NTR eQTL dataset: NESDA and NTR studies were approved by the Central Ethics Committee on Research Involving Human Subjects of the VU University Medical Center, Amsterdam (IRB number IRB-2991 under Federalwide Assurance 3703; IRB/institute codes, NESDA 03-183; NTR 03-180), and all subjects provided written informed consent. The sample used for eQTL analysis consisted of 5,071 subjects, 3,109 NTR (from 1,571 families: 614 dizygotic twin pairs, 1 monozygotic triplet, 668 monozygotic twin pairs, 394 siblings and 148 unrelated subjects) and 1,962 NESDA participants. The age of the participants ranged from 17 to 88 years (mean 38, SD 13) and 65% of the sample was female. Western Australia Pregnancy Study (RAINE): Recruitment of the Western Australian Pregnancy (RAINE) cohort has previously been described in detail (21–23). In brief, between 1989 and 1991, 2,900 pregnant women were recruited prior to 18-weeks gestation into a randomised controlled trial to evaluate the effects 23 of repeated ultrasound in pregnancy. Children have been comprehensively phenotyped from birth to 21 years of age (average ages of one, two, three, six, eight, ten, fourteen, seventeen and twenty-one) by trained members of the Raine research team. Most of the children are of Caucasian ethnicity. Data collection included questionnaires completed by the child’s primary carer and by the adolescent from age 14, physical assessments by trained assessors at all follow up years, and DNA collection from year 14 follow-up. The study was conducted with appropriate institutional ethics approval, and written informed consent was obtained from all mothers. Study individual genotype data was extracted from the genome-wide Illumina 660 Quad Array. TEENS of Attica: Genes and Environment Study (TEENAGE): The TEENAGE study is a cross-sectional study. The study target population comprised 857 adolescent students aged 13–15 years attending the first three classes of public secondary schools located in the wider Athens area of Attica. Prior to recruitment all study participants gave their verbal assent along with their parents’/guardians’ written consent forms. The study protocol was approved by the Institutional Review Board of Harokopio University and the Greek Ministry of Education, Lifelong Learning and Religious Affairs (24). Sexual maturity status was assessed by self-evaluation of the individual (25, 26) in the presence of the team’s paediatrician according to Tanner’s criteria (27) for breast, pubic hair and genital development. DNA samples of 707 study participants were genotyped using Illumina HumanOmniExpress BeadChips (Illumina, San Diego, CA, USA) at the Wellcome Trust Sanger Institute, Hinxton, UK. Genotyping and data quality control have been described previously (28). Genotypes were called using Illuminus algorithm (29) and SNPs were imputed using the program IMPUTE (30). Infancia y Medio Ambiente (Environment and childhood) Project (INMA): Population-based birth cohorts were established as part of the INMA – INfancia y Medio Ambiente [Environment and Childhood] Project in several regions of Spain following a common protocol. This analysis uses the INMA cohort of Menorca (Balearic Islands) established between 1997 and 1998. This project aims to study the associations between pre- and postnatal environmental exposures and growth, health, and development from early fetal life until adolescence and has been described previously in detail (31). Pregnant women were enrolled during the pregnancy at public primary health care centers or public hospitals. Detailed measurements were performed using physical examinations and biological samples were collected. Informed consent was obtained from all participants and the study was approved by the Hospital Ethics Committee. Whole blood or saliva DNA collected at age 4y was available for 432/482 (89%) children from Menorca. Samples from white European children were selected for the genome-wide genotyping using the HumanOmni1-Quad Beadchip (Illumina). This analysis uses data from 119 females from the cohort. Leipzig Childhood Cohort (LEIPZIG): A representative sample of children and adolescents from Central Germany was collected in a cross-sectional study from schools. Schools were selected to cover representative local areas within the city of Leipzig and its suburbs (hence social distribution) and covering different school types, hence levels of education. Physical examinations were performed by pediatricians. Height was measured using a standardized mobile digital stadiometer and weight by digital scale with an accuracy of 0.1 kg. The study was approved by the local ethics committee #054-2006. All guardians and children gave informed consent to the study and analyses. 24 Special Turku Coronary Risk Factor Intervention Project (STRIP): The STRIP trial is a prospective, randomized study that aims to prevent atherosclerosis beginning in infancy. The main objectives of STRIP have been to study the safety and effects of a low saturated fat diet in childhood. In brief, between February 1990 and June 1992, families of 6-month-old infants were recruited at well-baby clinics in Turku, Finland. At 7 months of age, 1,062 infants were randomly allocated to a dietary intervention (n=540) or control (n=522) group. The intervention group received individualized dietary and subsequently antismoking counseling at least biannually until 20 years of age. The STRIP study has several dimensions. First, the follow-up period of the intervention and control children and families is long, i.e. from 7 months of age through childhood and adolescence till early adulthood. The coronary risk factor profile exploration has been wide ranging (serum lipids and lipoprotein and apolipoproteins, glucose and insulin values, blood pressure, recurrent infections, oral health, etc.) and included also many measurable aspects of daily life (diet, physical activity, smoking, socioeconomic status, psychosocial well-being, etc.) have been explored. Vascular ultrasonic measurements were introduced at the age of 11 and repeated every two years thereafter. The core laboratory examinations have covered the key atherosclerosis risk factors and these values and aspects have been monitored in all STRIP children regularly and frequently, most of them annually. Pubertal status of the child was recorded from the age of 9 years onward. The physicians who examined the children were trained by an experienced pediatric endocrinologist to correctly stage pubertal development. The signs of puberty were recorded according to Tanner staging. Breast tissue diameter and testicular length were measured with a ruler, and pubic hair development was estimated visually. The respective pubertal stages (M1 through M5 and P1 through P5 in girls; G1 through G5 and P1 through P5 in boys) were recorded according to well established criteria. M2 stood for breast budding; G2 indicated testicular length of 20 mm (corresponding volume 3 mL). The study was approved by the Joint Commission on Ethics of the Turku University and the Turku University Central Hospital. Written informed consent was obtained from the parents in the beginning of the study and from the adolescents at 15 years of age. The Joint Commission on Ethics of the Turku University and the Turku University Central Hospital approved the STRIP study. Informed consent was obtained from the parents of the children at the beginning of the trial. References 1. Magi, R., Lindgren, C.M. and Morris, A.P. (2010) Meta-analysis of sex-specific genome-wide association studies. Genet. Epidemiol., 34, 846–853. 2. Mägi, R. and Morris, A.P. (2010) GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics, 11, 288. 3. Boyle, A.P., Hong, E.L., Hariharan, M., Cheng, Y., Schaub, M.A., Kasowski, M., Karczewski, K.J., Park, J., Hitz, B.C., Weng, S., et al. (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res., 22, 1790–1797. 25 4. Speliotes, E.K., Willer, C.J., Berndt, S.I., Monda, K.L., Thorleifsson, G., Jackson, A.U., Lango Allen, H., Lindgren, C.M., Luan, J., Mägi, R., et al. (2010) Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet., 42, 937–948. 5. Elks, C.E., Perry, J.R.B., Sulem, P., Chasman, D.I., Franceschini, N., He, C., Lunetta, K.L., Visser, J.A., Byrne, E.M., Cousminer, D.L., et al. (2010) Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat. Genet., 42, 1077–1085. 6. Cousminer, D.L., Berry, D.J., Timpson, N.J., Ang, W., Thiering, E., Byrne, E.M., Taal, H.R., Huikari, V., Bradfield, J.P., Kerkhof, M., et al. (2013) Genome-wide association and longitudinal analyses reveal genetic loci linking pubertal height growth, pubertal timing and childhood adiposity. Hum. Mol. Genet., 10.1093/hmg/ddt104. 7. Fernández-Rhodes, L., Demerath, E.W., Cousminer, D.L., Tao, R., Dreyfus, J.G., Esko, T., Smith, A.V., Gudnason, V., Harris, T.B., Launer, L., et al. (2013) Association of Adiposity Genetic Variants With Menarche Timing in 92,105 Women of European Descent. Am. J. Epidemiol., 10.1093/aje/kws473. 8. Demerath, E.W., Liu, C.-T., Franceschini, N., Chen, G., Palmer, J.R., Smith, E.N., Chen, C.T.L., Ambrosone, C.B., Arnold, A.M., Bandera, E.V., et al. (2013) Genome-wide association study of age at menarche in African-American women. Hum. Mol. Genet., 10.1093/hmg/ddt181. 9. Bradfield, J.P., Taal, H.R., Timpson, N.J., Scherag, A., Lecoeur, C., Warrington, N.M., Hypponen, E., Holst, C., Valcarcel, B., Thiering, E., et al. (2012) A genome-wide association meta-analysis identifies new childhood obesity loci. Nat. Genet., 44, 526–531. 10. Segrè, A.V., DIAGRAM Consortium, MAGIC investigators, Groop, L., Mootha, V.K., Daly, M.J. and Altshuler, D. (2010) Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet., 6. 11. Boyd, A., Golding, J., Macleod, J., Lawlor, D.A., Fraser, A., Henderson, J., Molloy, L., Ness, A., Ring, S. and Smith, G.D. (2012) Cohort Profile: The ‘Children of the 90s’—the index offspring of the Avon Longitudinal Study of Parents and Children. Int. J. Epidemiol., 10.1093/ije/dys064. 12. Li, Y., Willer, C.J., Ding, J., Scheet, P. and Abecasis, G.R. (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol., 34, 816–834. 13. Howie, B., Fuchsberger, C., Stephens, M., Marchini, J. and Abecasis, G.R. (2012) Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet., 44, 955– 959. 14. International Multiple Sclerosis Genetics Consortium, Wellcome Trust Case Control Consortium 2, Sawcer, S., Hellenthal, G., Pirinen, M., Spencer, C.C.A., Patsopoulos, N.A., Moutsianas, L., Dilthey, A., Su, Z., et al. (2011) Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature, 476, 214–219. 15. Barrett, J.C., Clayton, D.G., Concannon, P., Akolkar, B., Cooper, J.D., Erlich, H.A., Julier, C., Morahan, G., Nerup, J., Nierras, C., et al. (2009) Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet., 41, 703–707. 26 16. Heinonen, I., Helajarvi, H., Pahkala, K., Heinonen, O.J., Hirvensalo, M., Palve, K., Tammelin, T., Yang, X., Juonala, M., Mikkila, V., et al. (2013) Sedentary behaviours and obesity in adults: the Cardiovascular Risk in Young Finns Study. BMJ Open, 3. 17. Van Beijsterveldt, C.E.M., Groen-Blokhuis, M., Hottenga, J.J., Franić, S., Hudziak, J.J., Lamb, D., Huppertz, C., de Zeeuw, E., Nivard, M., Schutte, N., et al. (2013) The Young Netherlands Twin Register (YNTR): longitudinal twin and family studies in over 70,000 children. Twin Res. Hum. Genet. Off. J. Int. Soc. Twin Stud., 16, 252–267. 18. Van Soelen, I.L.C., Brouwer, R.M., van Baal, G.C.M., Schnack, H.G., Peper, J.S., Chen, L., Kahn, R.S., Boomsma, D.I. and Hulshoff Pol, H.E. (2013) Heritability of volumetric brain changes and height in children entering puberty. Hum. Brain Mapp., 34, 713–725. 19. Hoekstra, R.A., Bartels, M. and Boomsma, D.I. (2006) Heritability of testosterone levels in 12-year-old twins and its relation to pubertal development. Twin Res. Hum. Genet. Off. J. Int. Soc. Twin Stud., 9, 558–565. 20. Scheet, P., Ehli, E.A., Xiao, X., van Beijsterveldt, C.E.M., Abdellaoui, A., Althoff, R.R., Hottenga, J.J., Willemsen, G., Nelson, K.A., Huizenga, P.E., et al. (2012) Twins, tissue, and time: an assessment of SNPs and CNVs. Twin Res. Hum. Genet. Off. J. Int. Soc. Twin Stud., 15, 737–745. 21. Newnham, J.P., Sharon, S.F., Michael, C.A., Stanley, F.J. and Landau, L.I. (1994) [Effects of frequent ultrasound during pregnancy: a randomised controlled trial]. Jordemodern, 107, 83–86. 22. Williams, L.A., Evans, S.F. and Newnham, J.P. (1997) Prospective cohort study of factors influencing the relative weights of the placenta and the newborn infant. BMJ, 314, 1864–1868. 23. Evans, S., Newnham, J., MacDonald, W. and Hall, C. (1996) Characterisation of the possible effect on birthweight following frequent prenatal ultrasound examinations. Early Hum. Dev., 45, 203–214. 24. Ntalla, I., Giannakopoulou, M., Vlachou, P., Giannitsopoulou, K., Gkesou, V., Makridi, C., Marougka, M., Mikou, G., Ntaoutidou, K., Prountzou, E., et al. (2013) Body composition and eating behaviours in relation to dieting involvement in a sample of urban Greek adolescents from the TEENAGE (TEENs of Attica: Genes & Environment) study. Public Health Nutr., 10.1017/S1368980013000074. 25. Duke, P.M., Litt, I.F. and Gross, R.T. (1980) Adolescents’ self-assessment of sexual maturation. Pediatrics, 66, 918–920. 26. Bonat, S., Pathomvanich, A., Keil, M.F., Field, A.E. and Yanovski, J.A. (2002) Self-assessment of pubertal stage in overweight children. Pediatrics, 110, 743–747. 27. Garn, S.M. (1956) Growth at adolescence. By J. M. Tanner. Pp. vii + 212. Blackwell Scientific Publications, Oxford. Publisher simultaneously by Charles C Thomas and the Ryerson Press. 1955. Am. J. Phys. Anthropol., 14, 120–122. 28. Ntalla, I., Panoutsopoulou, K., Vlachou, P., Southam, L., William Rayner, N., Zeggini, E. and Dedoussis, G.V. (2013) Replication of established common genetic variants for adult BMI and childhood obesity in Greek adolescents: the TEENAGE study. Ann. Hum. Genet., 77, 268–274. 27 29. Teo, Y.Y., Inouye, M., Small, K.S., Gwilliam, R., Deloukas, P., Kwiatkowski, D.P. and Clark, T.G. (2007) A genotype calling algorithm for the Illumina BeadArray platform. Bioinforma. Oxf. Engl., 23, 2741–2746. 30. Marchini, J., Howie, B., Myers, S., McVean, G. and Donnelly, P. (2007) A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet., 39, 906–913. 31. Guxens, M., Ballester, F., Espada, M., Fernández, M.F., Grimalt, J.O., Ibarluzea, J., Olea, N., Rebagliato, M., Tardón, A., Torrent, M., et al. (2012) Cohort Profile: the INMA--INfancia y Medio Ambiente-(Environment and Childhood) Project. Int. J. Epidemiol., 41, 930–940. Consortia 1. The Early Growth Genetics (EGG) Consortium Linda S Adair, Wei Ang, Mustafa Atalay, Toos van Beijsterveldt, Nienke Bergen, Kelly Benke, Diane J Berry, Dorret I Boomsma, Jonathan P Bradfield, Pimphen Charoen, Lachlan Coin, Cyrus Cooper, Diana L Cousminer, Shikta Das, Oliver S P Davis, George V Dedoussis, Paul Elliott, Xavier Estivill, David M Evans, Bjarke Feenstra, Claudia Flexeder, Tim Frayling, Rachel M Freathy, Romy Gaillard, Frank Geller, Matthew Gillman, Struan F A Grant, Maria Groen-Blokhuis, Liang-Kee Goh, Mònica Guxens, Hakon Hakonarson, Andrew T Hattersley, Claire M A Haworth, Dexter Hadley, Johannes Hedebrand, Joachim Heinrich, Anke Hinney, Joel N Hirschhorn, Berthold Hocher, John W Holloway, Claus Holst, Jouke Jan Hottenga, Momoko Horikoshi, Ville Huikari, Elina Hypponen, Carmen Iñiguez, Vincent WV Jaddoe, Marjo-Riitta Jarvelin, Marika Kaakinen, Tuomas O Kilpeläinen, Mirna Kirin, Matthew Kowgier, Hanna-Maaria Lakka, Timo A Lakka, Leslie A Lange, Debbie A Lawlor, Terho Lehtimäki, Alex Lewin, Cecilia Lindgren, Virpi Lindi, Reedik Maggi, Julie Marsh, Mark I McCarthy, Mads Melbye, Christel Middeldorp, Iona Millwood, Karen L Mohlke, Dennis O Mook-Kanamori, Jeffrey C Murray, Michel Nivard, Ellen Aagaard Nohr, Ioanna Ntalla, Emily Oken, Ken K Ong, Paul F O’Reilly, Lyle J Palmer, Kalliope Panoutsopoulou, Jennifer Pararajasingham, Ewan R Pearson, Craig E Pennell, Chris Power, Thomas S Price, Inga Prokopenko, Olli T Raitakari, Alina Rodriguez, Rany M Salem, Seang-Mei Saw, Andre Scherag, Sylvain Sebert, Niina Siitonen, Olli Simell, Thorkild I A Sørensen, Ulla Sovio, Beate St Pourcain, David P Strachan, Jordi Sunyer, H Rob Taal, Yik-Ying Teo, Elisabeth Thiering, Carla Tiesler, Nicholas J Timpson, Andre G Uitterlinden, Beatriz Valcárcel, Nicole M Warrington, Scott White, Elisabeth Widén, Gonneke Willemsen, James F Wilson, Hanieh Yaghootkar & Eleftheria Zeggini. 2. The ReproGen Consortium Cathy E Elks, John R B Perry, Patrick Sulem, Daniel I Chasman, Nora Franceschini, Chunyan He, Kathryn L Lunetta, Jenny A Visser, Enda M Byrne, Diana L Cousminer, Daniel F Gudbjartsson, Tõnu Esko, Bjarke Feenstra, Jouke-Jan Hottenga, Daniel L Koller, Zoltán Kutalik, Peng Lin, Massimo Mangino, Mara Marongiu, Patrick F McArdle, Albert V Smith, Lisette Stolk, Sophie H van Wingerden, Jing Hua Zhao, Eva Albrecht, Tanguy Corre, Erik Ingelsson, Caroline Hayward, Patrik K E Magnusson, Erin N Smith, Shelia Ulivi, Nicole M Warrington, Lina Zgaga, Helen Alavere, Najaf Amin, Thor Aspelund, Stefania Bandinelli, Inês Barroso, Gerald S Berenson, Sven Bergmann, Hannah Blackburn, Eric Boerwinkle, Julie E Buring, Fabio Busonero, Harry Campbell, Stephen J Chanock, Wei Chen, Marilyn C Cornelis, David Couper, Andrea D Coviello, Pio d'Adamo, Ulf de Faire, Eco J C de Geus, Panos Deloukas, Angela Döring, George Davey Smith, Douglas F Easton, Gudny Eiriksdottir, Valur Emilsson, Johan Eriksson, Luigi Ferrucci, Aaron R Folsom, Tatiana Foroud, Melissa Garcia, Paolo Gasparini, Frank Geller, Christian Gieger, The GIANT Consortium, Vilmundur Gudnason, Per Hall, Susan E Hankinson, Liana Ferreli, Andrew C 28 Heath, Dena G Hernandez, Albert Hofman, Frank B Hu, Thomas Illig, Marjo-Riitta Järvelin, Andrew D Johnson, David Karasik, Kay-Tee Khaw, Douglas P Kiel, Tuomas O Kilpeläinen, Ivana Kolcic, Peter Kraft, Lenore J Launer, Joop S E Laven, Shengxu Li, Jianjun Liu, Daniel Levy, Nicholas G Martin, Wendy L McArdle, Mads Melbye, Vincent Mooser, Jeffrey C Murray, Sarah S Murray, Michael A Nalls, Pau Navarro, Mari Nelis, Andrew R Ness, Kate Northstone, Ben A Oostra, Munro Peacock, Lyle J Palmer, Aarno Palotie, Guillaume Paré, Alex N Parker, Nancy L Pedersen, Leena Peltonen, Craig E Pennell, Paul Pharoah, Ozren Polasek, Andrew S Plump, Anneli Pouta, Eleonora Porcu, Thorunn Rafnar, John P Rice, Susan M Ring, Fernando Rivadeneira, Igor Rudan, Cinzia Sala, Veikko Salomaa, Serena Sanna, David Schlessinger, Nicholas J Schork, Angelo Scuteri, Ayellet V Segrè, Alan R Shuldiner, Nicole Soranzo, Ulla Sovio, Sathanur R Srinivasan, David P Strachan, Mar-Liis Tammesoo, Emmi Tikkanen, Daniela Toniolo, Kim Tsui, Laufey Tryggvadottir, Jonathon Tyrer, Manuela Uda, Rob M van Dam, Joyce B J van Meurs, Peter Vollenweider, Gerard Waeber, Nicholas J Wareham, Dawn M Waterworth, Michael N Weedon, H Erich Wichmann, Gonneke Willemsen, James F Wilson, Alan F Wright, Lauren Young, Guangju Zhai, Wei Vivian Zhuang, Laura J Bierut, Dorret I Boomsma, Heather A Boyd, Laura Crisponi, Ellen W Demerath, Cornelia M van Duijn, Michael J Econs, Tamara B Harris, David J Hunter, Ruth J F Loos, Andres Metspalu, Grant W Montgomery, Paul M Ridker, Tim D Spector, Elizabeth A Streeten, Kari Stefansson, Unnur Thorsteinsdottir, André G Uitterlinden, Elisabeth Widen, Joanne M Murabito, Ken K Ong & Anna Murray. 29