[CLICK HERE AND TYPE TITLE]
advertisement
![[CLICK HERE AND TYPE TITLE]](http://s3.studylib.net/store/data/006920558_1-463cb03fce964e9240461fa313156bf5-768x994.png)
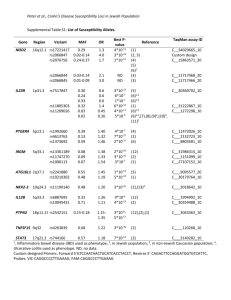
International Biometric Society A POWERFUL TWO-STAGE TEST FOR GENE-TO-GENE INTERACTIONS IN A CASE-CONTROL SETTING Jakub Pecanka (1), Marianne Jonker (2), Zoltan Bochdanovits (3), Aad van der Vaart (4) (1) Department of Mathematics, VU University, Amsterdam, the Netherlands (2) Department of Epidemiology and Biostatistics, VU Medical Center, the Netherlands (3) Department of Clinical Genetics, VU Medical Center, the Netherlands (4) Department of Mathematics, Leiden University, the Netherlands Functional gene-gene interaction (epistasis) is a biologically very plausible explanation for, part of, the “missing heritability" of complex traits. Although this possibility is widely acknowledged, a simple full screen for epistatic effects on a genome-wide scale is still considered unfeasible, because of serious loss of statistical power due to necessary multiple testing correction. A possible way to lessen the extent of this problem is to ascertain pairs of variants that are a priori more likely to be involved in functional gene-gene interactions and consider only these when testing for association with a complex phenotype. Here we present several procedures for detecting genome-wide significant gene-gene interactions that are all based on a chisquare-type pre-test to select pairs of loci that exhibit nonequilibrium two locus genotype frequencies in the (control) population, followed by a scoretype test for interaction that is adjusted for re-using data in the second step testing. In these procedures, the necessary multiple testing correction reduces to the number of tests in the second step, while the type I error control is correctly maintained. In the presentation the power of different procedures are compared for different interaction models. We show that a two step procedure has often superior power compared to the related one step interaction test with multiple testing correction for all pairs involved in the analysis. Since only the chisquare-type pre-test is performed for all pairs of loci, the procedure is computationally feasible and efficient while the reduction of the number of tests in the second step conveys increased overall power making this procedure suitable to analyze genome-wide data. International Biometric Conference, Florence, ITALY, 6 – 11 July 2014