1
Supporting Information
2
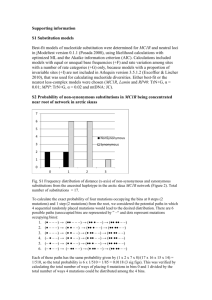
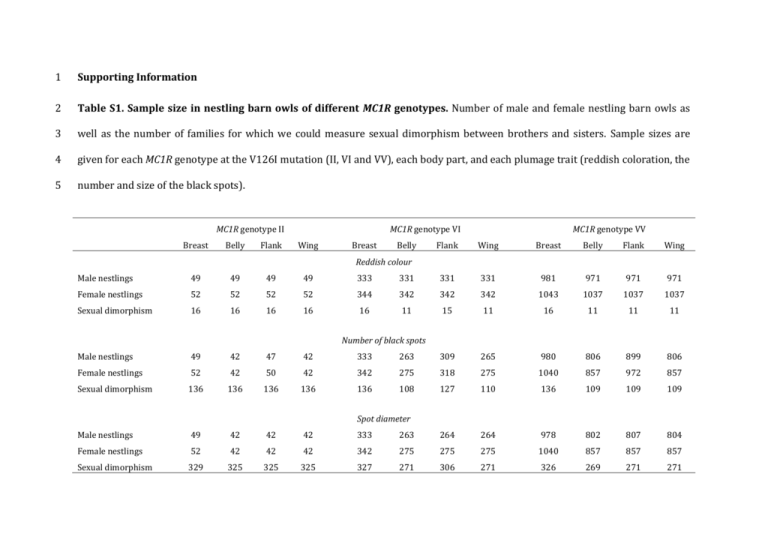
Table S1. Sample size in nestling barn owls of different MC1R genotypes. Number of male and female nestling barn owls as
3
well as the number of families for which we could measure sexual dimorphism between brothers and sisters. Sample sizes are
4
given for each MC1R genotype at the V126I mutation (II, VI and VV), each body part, and each plumage trait (reddish coloration, the
5
number and size of the black spots).
MC1R genotype II
Breast
Belly
Flank
MC1R genotype VI
Wing
Breast
Belly
MC1R genotype VV
Flank
Wing
Breast
Belly
Flank
Wing
Reddish colour
Male nestlings
49
49
49
49
333
331
331
331
981
971
971
971
Female nestlings
52
52
52
52
344
342
342
342
1043
1037
1037
1037
Sexual dimorphism
16
16
16
16
16
11
15
11
16
11
11
11
Number of black spots
Male nestlings
49
42
47
42
333
263
309
265
980
806
899
806
Female nestlings
52
42
50
42
342
275
318
275
1040
857
972
857
Sexual dimorphism
136
136
136
136
136
108
127
110
136
109
109
109
Spot diameter
Male nestlings
49
42
42
42
333
263
264
264
978
802
807
804
Female nestlings
52
42
42
42
342
275
275
275
1040
857
857
857
Sexual dimorphism
329
325
325
325
327
271
306
271
326
269
271
271
6
7
Table S2. Number of male and female nestling barn owls sampled between 1996
8
and 2013.
Year
Male nestlings
Female nestlings
Sum
1996
94
114
208
1997
48
61
109
1998
90
91
181
1999
65
66
131
2000
12
9
21
2001
85
96
181
2002
131
121
252
2003
88
64
152
2004
59
88
147
2005
90
85
175
2006
28
35
63
2007
148
120
268
2008
97
104
201
2009
21
32
53
2010
86
82
168
2011
66
91
157
2012
147
170
317
2013
9
10
19
9
Table S3. Number of barn owls for which we measured plumage traits over several years. Sample sizes are given for each
10
MC1R genotype at the V126I mutation (II, VI and VV), each body part, sex and plumage trait. For the statistical analysis on age-
11
related colour changes, II individuals were excluded given the low sample size.
MC1R genotype II
Breast
Belly
Flank
MC1R genotype VI
Wing
Breast
Belly
Flank
MC1R genotype VV
Wing
Breast
Belly
Flank
Wing
Reddish colour
Adult males
10
10
10
10
91
91
91
91
234
234
234
234
Adult females
11
11
11
11
120
120
120
120
317
317
317
317
Number of black spots
Adult males
10
10
10
10
91
88
88
88
234
226
226
226
Adult females
11
11
11
11
120
111
111
111
317
300
300
300
Spot diameter
Adult males
10
10
10
10
91
76
76
76
234
199
200
200
Adult females
11
6
6
6
120
94
94
94
317
258
258
258
12
Table S4. Sequences of the Primers used in this study.
Primer Name
Sequence (from 5' to 3')
a) Primers designed on Gallus
MC1R_43Fw
AACGCCAGTGAGGGCAACCA
MC1R_944Rev
TACCAGGAGCACAGCACCACCT
b) Genome walking primers
MC1R_660fw
CATCCTCCTGGGCGTCTTCTTCATCT
MC1R_775fw
TCCACATCCTCATCATCTGCAACTCGG
c) Race primers
MC1RTa_134Rev
AGGAAGAGCCCGTTGGGGATGT
MC1RTa_228Rev
CAGATGAAGTAGTACGTGGGCGAGTG
d) Sequencing specific primers
13
14
MC1R_-34fw
GGGACCCCGGGGTTGAGGCG
MC1R_568rev
GGCAGAGGAGGATGGCGTTGTTGCG
MC1R_404fw
TCATCGCCGTGGACCGCTACATCACCA
MC1R_969rev
GCGTTAACCCGCGTCCCGCTGC
e) Allelic discrimination assay primers
MC1R_198fw
MC1R_453Rev
CCTGCACTCGCCCACGTACTACTTC
GTGGTAGCGCAGGGCGTAGAAGAT
V126I_fw
CATGGACAACGTCATCGA
V126I_rev
GCGTAGAAGATGGTGATGTA
V126I_wt_Fam-BHQ1
TGCAGCTCCGTCGTGTCCTC
V126I_mut_ATTO550-BHQ2
TGCAGCTCCATCGTGTCCTC
15
Supplementary Materials and Methods
16
Genome walking were used to identify the 3’ end of the MC1R coding sequence.
17
According to the GenomeWalker universal kit (Clontech, Takara Bio Europe/Clontech,
18
Saint-Germain-en-Laye, France), the primers MC1R_660fw was used in combination
19
with AP1, while primers MC1R_775fw was used in combination with AP2 primer from
20
the universal kit during the initial and nested PCR, respectively. Cycling conditions and
21
polymerase were those recommended by the kit.
22
For the Race, total RNAs were extracted from growing feather bases using RNeasy mini
23
kit (Qiagen, Hombrechtikon, Switzerland) and 2.5 g of total RNA was reverse
24
transcribed with oligodT primer and Superscript III and proceeded to the RACE assay
25
following the GeneRacer kit protocol (Life Technologies, Zug, Switzerland) and cloned
26
and sequenced as described in Materials and Methods. 500 nM MC1RTa228Rev was
27
used in combination with 500 nM GeneRacer-5’ to the first amplification of cDNA
28
prepared as described in the kit with 200M dNTPs, 1x Kapa buffer A, 1x Kapa
29
Enhancer, 1U Kapa Robust 2G (Labgene Scientific, Châtel-St-Denis, Switzerland) in 50 l
30
with a first denaturation at 95°C for 5 min, a touchdown cycling with 95°C 25 sec, a
31
decrease of 1°C at each cycle for 10 cycles for the annealing starting at 70°C 30 sec and
32
72°C 1 min, followed by 32 cycles with 95°C 25sec, 60°C 30 sec, 72°C 1 min and a final
33
elongation at 72°C for 10 min. The nested PCR was then conducted in 50 l with 1/50 of
34
the product of the first PCR with 500 nM GeneRacer-5’-nested, 500 nM MC1RTa134Rev
35
and the Kapa Robust 2G (Labgene) as above with the following conditions: 95°C 5min,
36
35 cycles at 95°C 25 sec, 59°C 30 sec, 72°C 1min and 72°C 10 min. The PCR products
37
were then gel purified on 1% agarose in 1xTBE with the Minelute Gel extraction kit
38
(Qiagen, Hombrechtikon, Switzerland), cloned and sequenced as described previously.
39
For the allelic discrimination (AD) assay, pre-amplification PCRs were performed using
40
exactly 20 ng of DNA sample, 1x Q-solution (Qiagen, Hombrechtikon, Switzerland),
41
200µM MC1R_-198fw and MC1R_453rev primers, 0.2 U of Taq (Qiagen, Hombrechtikon,
42
Switzerland) into a final volume of 20µl with the following cycle conditons: 95 °C for 5
43
min, 34 cycles at 94 °C for 30 sec, 63 °C for 30 sec, and 72 °C for 30 sec, and final
44
extension at 72°C for 10 min. As initial DNA concentration is critical for AD assays,
45
relative quantity of the PCR products were compared using a 2% agarose gel and
46
adjusted among each other and then diluted 100 times before the AD assay. AD assays
47
were run in an ABI 7500 qPCR machine (Life Technologies, Zug, Switzerland). Each
48
qPCR plate contained three positive controls (corresponding to each genotype) and at
49
least two negative controls. The qPCR MasterMix Plus Low ROX (Eurogentec, Liège,
50
Belgium) was used with an annealing temperature of 57°C for 60 sec in a final volume of
51
24 µL with 300 nM of V126I_fw and V126I_rev, 100 nM of V126I_wt_Fam-BHQ1
52
(Microsynth, Balgach, Switzerland), 250 nM V126I_mut_ATTO550-BHQ2 (Microsynth,
53
Balgach, Switzerland) and 2 µl of diluted DNA.
54
55
Supplementary results:
56
Relative impact of MC1R on plumage traits in the barn owl
57
To statistically test the relative impact of MC1R on the three plumage traits, we
58
performed linear mixed models on standardized plumage trait values for each body part
59
and using nestling, maternal and paternal identities as random factors and sex, MC1R,
60
type of plumage trait (reddish coloration, number of spots, spot diameter), and all their
61
interaction as independent variables. On the breast, the impact of MC1R was stronger on
62
reddish coloration than number of spots (interaction MC1R plumage trait: F2,2793 =
63
309.82, P < 0.0001), on reddish coloration than spot diameter (interaction MC1R
64
plumage trait: F2,2788 = 228.18, P < 0.0001) and on spot diameter than number of spots
65
(interaction MC1R plumage trait: F2,2788 = 14.50, P < 0.0001). We obtained similar
66
results for the belly, flank, and the underside of the wings (all P-values < 0.04) except
67
that, on the flank, the impact of MC1R on the number and size of spots was not
68
significantly different (interaction: F2,2458 = 1.88, P = 0.15).
69