projects for 2016 - IMB Postgraduate website
advertisement
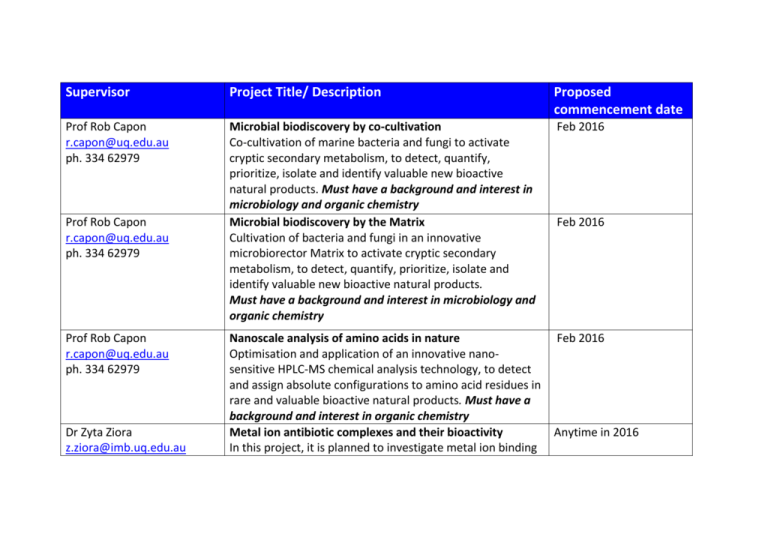
Supervisor Project Title/ Description Proposed commencement date Prof Rob Capon r.capon@uq.edu.au ph. 334 62979 Microbial biodiscovery by co-cultivation Co-cultivation of marine bacteria and fungi to activate cryptic secondary metabolism, to detect, quantify, prioritize, isolate and identify valuable new bioactive natural products. Must have a background and interest in microbiology and organic chemistry Microbial biodiscovery by the Matrix Cultivation of bacteria and fungi in an innovative microbiorector Matrix to activate cryptic secondary metabolism, to detect, quantify, prioritize, isolate and identify valuable new bioactive natural products. Must have a background and interest in microbiology and organic chemistry Feb 2016 Nanoscale analysis of amino acids in nature Optimisation and application of an innovative nanosensitive HPLC-MS chemical analysis technology, to detect and assign absolute configurations to amino acid residues in rare and valuable bioactive natural products. Must have a background and interest in organic chemistry Metal ion antibiotic complexes and their bioactivity In this project, it is planned to investigate metal ion binding Feb 2016 Prof Rob Capon r.capon@uq.edu.au ph. 334 62979 Prof Rob Capon r.capon@uq.edu.au ph. 334 62979 Dr Zyta Ziora z.ziora@imb.uq.edu.au Feb 2016 Anytime in 2016 ph. 334 62067 using titration experiments by isothermal titration calorimetry (ITC) and NMR. The metal ion-antibiotic complexes will be submitted for MIC assays to examine the metal ions role in applied biological systems. Dr Zyta Ziora Antibiotic release from magnetic nanoparticles for z.ziora@imb.uq.edu.au improved biopotency ph. 334 62067 Antibiotic-peptide-linker-antibody construct will recognise bacteria maturation. The enzyme from damaged membrane will be exposed to the surface of bacteria and it will recognise the peptidic sequence being in the linker; the peptide will be enzymatically cleaved releasing free antibiotic and causing their decease. Dr Mark Butler Identification of biologically active natural products m.butler5@uq.edu.au As part of an ongoing efforts to identify new anti-infective ph. 334 62992 leads, we have identified extracts with antifungal and antibacterial activity. Bioassay-guided isolation will be used to identify the active components and their structures determined using spectroscopic techniques that include NMR and MS. Prof Matt Cooper / Wanida Synthesis of novel bacitracin antibiotic probes Phetsang This project will prepare Bacitracin analogues with azide Infogroups at these positions, as useful starting materials for coopergroup@imb.uq.edu.au generating both fluorescent probes and hybrid antibiotics ph. 334 62044 visa ‘click’ chemistry. The fluorescent probe(s) will be used Anytime in 2016 Anytime in 2016 Anytime in 2016 to investigate the Bacitracin A molecular targets. Prof Matt Cooper / Angie Synthesis of novel antibiotics to combat drug resistant Anytime in 2016 Jarrad Clostridium difficile InfoThe aim of this project is to prepare novel next generation coopergroup@imb.uq.edu.au antibiotics that can overcome deficiencies in the current Ph. 334 62044 treatment options. Compounds prepared by the student will contribute to an exciting multidisciplinary, drug discovery program to combat this debilitating human disease. Prof Matt Cooper / Dr Karl Hit validation of novel antimicrobial molecules Anytime in 2016 Hansford This chemistry based project aims synthesis analogues of hit Infomolecules as part of the hit validation process to obtain coopergroup@imb.uq.edu.au structure-activity information. Ph. 334 62044 Dr Cas Simons c.simons@imb.uq.edu.au ph. 334 62080 Characterisation of Zebrafish models of white matter Anytime in 2016 disorders Leukodystrophies are inherited diseases that affect myelin in the central nervous system. These diseases usually present early in life, are progressive and incurable. Through whole genome of families affected by rare and undiagnosed leukodystrophies we have identified mutations in several novel candidate genes. This project revolves around characterising the phenotype a zebrafish knock out model for one of these genes and will likely involve basic molecular A/Prof Rohan Teasdale r.teasdale@uq.edu.au ph. 334 62056 A/Prof Rohan Teasdale r.teasdale@uq.edu.au ph. 334 62056 Prof Mark Smythe / Dr Christina Kulis c.kulis@imb.uq.edu.au ph. 334 62368 biology (cloning, in vitro transcription), zebrafish breading, RNA in situ hybridization, immunofluorescence, qPCR and imaging of live/fixed zebrafish with fluorescent transgenic reporters. Characterisation of the mammalian endosomal protein Anytime in 2016 complex called the retromer Retromer is a central regulator of early endosome protein trafficking that has recently been implicated in the progressive neurological disorders such as Alzheimer’s and Parkinson’s disease. This project will examine the properties of retromer to determine the molecular mechanisms underlying these disease states. Defining the Endosomal Network Anytime in 2016 The endo-lysosomal system of mammalian cells incorporates highly dynamic endocytic membrane transport pathways that are fundamental for many cellular processes. Understanding how the mammalian endosomal system functions firstly requires its composition to be defined. This project will combine bioinformatics with classic cell biology techniques to help define this important biological system. Development of New Spin Label Probes Anytime in 2016 Electron paramagnetic resonance (EPR) spectroscopy is a powerful technique for studying the structure and dynamics of proteins. However its effectiveness is limited by the spin Prof Mark Smythe / Dr Christina Kulis c.kulis@imb.uq.edu.au ph. 334 62368 Dr Michele Bastiani m.bastiani@uq.edu.au Ph. 334 62335 labels employed, which are either too flexible or bulky and do not resemble natural amino acids. This project is focused on the development and synthesis of amino acid based spin label probes to improve EPR measurements in biological systems Development of LPGDS Inhibitors for Inflammation This project is focused on the development of small molecule inhibitors to modulate the lipocalin prostaglandin D2 synthase (LPGD2S) enzyme. This project will involve the chemical synthesis and biological activity of these compounds. How does the extracellular environment shape the plasma membrane Mechanical cues can impact cellular behaviour affecting the cell’s morphology and fate. This project will make use of synthetic smart surfaces to investigate how caveolae, invaginations of the plasma membrane which respond to membrane tension and stretch, are affected by changes to the mechanical properties of the extracellular environment. These studies will be a collaborative project between the Institute of Molecular Bioscience (IMB) and the Australian Institute for Bioengineering and Nanotechnology (AIBN), presenting the opportunity to work on different cellular biology techniques with emphasis on high-resolution Anytime in 2016 Anytime in 2016 microscopy. A/Prof Lachlan Coin / Dr Analysing bacterial antibiotics resistance with real-time Anytime in 2016 Minh Duc Cao MinION sequencing m.cao1@uq.edu.au The latest high-throughput sequencing technology, the ph. 334 62178 Oxford Nanopore MinION platform, presents many innovative features opening up potential for many applications not previously possible. Among these, the ability to sequence in real-time provides a unique opportunity for many time-critical applications. This project will develop computational methods to take advantages of the ability to sequence long reads in real-time of this technology, and to overcome its high error rates. These methods will be applied to characterise bacterial antibiotics resistance genomics from clinical collected samples, thereby identify genotypes associated with the development of antimicrobial resistance. Suitable for a student with a background in computational and a desire to work at the interface of computing and biology. A/Prof Lachlan Coin / Dr Alan The rapid identification of antibiotic resistant bacteria Anytime in 2016 Robertson from clinical samples with MinION a.robertson@imb.uq.edu.au With the rise of antibiotic resistant microbes, the need to ph. 334 62178 quickly identify the source of an infection and prescribe the correct treatment is essential. The current method used to Dr Markus Muttenthaler m.muttenthaler@uq.edu.au ph. 334 62985 identify the bacterial species present in a sample takes upwards of 48 hours, a time-scale too long for many patients. We have recently used minion, a novel next generation sequencing platform (Oxford Nanopore) to identify the species of bacteria in a mixed sample and characterise their antibiotic resistance profile. This project will apply our approach to clinical samples and investigate the capacity of our tool to identify specific species and strains of bacteria as well as report the presence of antibiotic resistance phenotypes. Ideal for a student with wet-lab experience. Endosomal escape for quantum dots using cell-penetrating Anytime in 2016 peptides Cell-penetrating peptides (CPPs) have become a mainstay technology for facilitating the delivery of a wide variety of nanomaterials to cells and tissues. Currently, the library of CPPs to choose from is still limited, with the HIV TAT-derived motif still being the most used. In this project we want to synthesise and characterise novel CPP motifs for their ability to facilitate delivery of luminescent semiconductor quantum dots (QDs) in a model cell culture system. The synthesis will be carried out using solid-phase peptide synthesis. Potential applications include diagnostic tools, selective cell labelling, and therapeutic applications. Dr Markus Muttenthaler m.muttenthaler@uq.edu.au ph. 334 62985 Dr Markus Muttenthaler m.muttenthaler@uq.edu.au ph. 334 62985 Towards a complete molecular toolbox for the Oxytocin Anytime in 2016 and Vasopressin receptors The oxytocin (OT)/vasopressin (VP) G protein coupled receptors are part of a 600-million-year-old signalling system responsible for reproduction, water balance and complex social behaviour. They form viable yet underexplored targets for high-profile indications such as autism, schizophrenia, stress, depression, anxiety, cancer and pain. Therapeutic development has been hampered by a lack of receptor subtype-selective ligands suitable for rodent and human studies. Understanding the structureactivity relationship of OT/VP is important for the design of better probes and drug candidates. In this project we want to probe the highly conserved proline position of OT/VP to gauge if it can be used to tune selectivity and potency. The ligands will be synthesised by solid-phase peptide synthesis. Total chemical synthesis of a trefoil factor peptide Anytime in 2016 Trefoil factor (TFF) peptides are secreted into the gut as part of the inflammatory response and are responsible for gut protection and gastrointestinal wound healing. Their compact structure makes TFFs extremely stable to proteolytic degradation and enables them to function in the hostile environment of the gastrointestinal tract. No receptor has been linked to the signalling of these peptides and understanding the mechanism of action is of fundamental importance for this class of molecules. Achieving the total chemical synthesis (using solid-phase peptide synthesis) would allow us to produce enhanced probes to study the mechanism of action in greater detail.