Genetics and Probability
advertisement

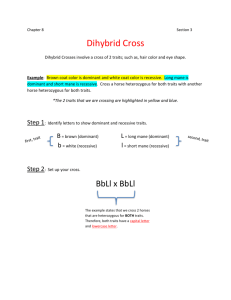
Classic Genetics AP Biology Genetics is about using the seen, the phenotype, to infer the unseen, the genotype. Scientists make predictions about modes of inheritance based on these inferences, and then compare the actual results with these expectations. Although at a glance experimental results may seem to support the prediction, best practice is to check the validity of data using a statistical test called Chi-square. Chi-square evaluates a hypothesis by determining whether or not the differences between observed and predicted results are due to chance or if they in fact represent a real difference. In statistics a difference is ‘significant’ if there is less than a 5% probability that the variation is due to random chance; if so, the hypothesis is rejected. Gregor Mendel initially considered a single trait of the pea at a time and from that deduced the Law of Dominance. Later, he investigated whether the inheritance of one trait influenced another, what he called the law of Independent Assortment. This involved predicting probabilities for multiple traits – for example, pea plants that are tall and produce yellow seeds. Multi-trait crosses have exponentially more outcomes, making a Punnett unwieldy; a better strategy is to determine the probability of each trait, and then multiply those probabilities. The Fast Plants seeds you grew last semester were dihybrid for leaf color and stem pigmentation. Later, Thomas Morgan replicated Mendel’s experiments using fruit flies. Morgan found that the flies’ traits were not inherited independent of one another when the genes were linked, or found on the same chromosome. The relative distance between two genes can be calculated from the offspring frequencies and a gene ‘map’ generated. The farther apart the genes are, the greater the likelihood crossing over will separate them. A. MENDEL’S PEA DATA Mendel first ran test crosses – pure dominant parent plants crossed with pure recessive. Then he crossed the results of the first generation (f1) with the same. 1. Calculate the total number of plants Mendel observed for any two traits. Using that, determine the predicted (expected) results for each trait. Recall that for traits such as these – only 2 alleles, one dominant, one recessive – the dominant phenotype is expected 75% of the time, the recessive 25%. Verify results with a partner. Record in Table 1. Parental traits Dominant Round seeds Purple flowers Inflated pods Green pods Axial flowers Tall stem Recessive Wrinkled White Constricted Yellow Terminal Dwarf Actual Dominant Actual Recessive 5474 705 882 428 651 787 1850 224 299 152 207 277 2. Since data almost always varies from the expected, we need to do a statistical test to see if the difference is significant – if it is, we would have to reject the hypothesis. We do this using the chi-square analysis, which is essentially an error analysis. Using the formula below, compare your actual results with the predicted. Record all calculations. Complete Table 1. (Actual dominant – expected dominant)2 Chi = ----------------------------------------------expected dominant 2 3. + (actual recessive – expected rec. )2 ------------------------------------expected recessive Look up your chi-square result in Table 2 as follows: • A chi-square result equal to no more than the 5% value means the observed differences between data and hypothesis are expected to occur due to chance alone at least 5% of the time. The closer the actual and expected are to each other, the greater likelihood the differences would be expected. If so, the hypothesis is accepted. In contrast, a chi-square larger than the value in the 5% column means the observed results are so different from the predicted we must conclude the prediction is wrong. • The degrees of freedom is one less than the number of groups. In this exercise there are two phenotypes (groups), therefore 1 degree of freedom (df). Find the values your chi square result falls between for 1 degree of freedom. Are your predictions supported by the data? Determine the nearest percent probability the difference between your hypothesis and results is due to chance. The higher this % the more confident we are in our hypothesis, but scientists accept any chi-square value that meets the 5% threshold. TABLE 2 – CHI2 RESULTS 95% 90% 80% 70% 50% 30% 20% 10% 5% df 1 0.004 0.02 0.064 0.15 0.46 1.07 1.64 2.71 3.84 2 0.10 0.21 0.45 0.71 1.39 2.41 3.22 4.60 5.99 3 0.35 0.58 1.00 1.42 2.37 3.66 4.64 6.25 7.82 B. FAST PLANT DIHYBRID DATA Carolina Biological, who developed the seeds, hypothesizes that green leaves are dominant to yellow, and anthocyanin pigment (creating purple stems) is dominant to no pigment (green stems). The f1 seed I purchase is heterozygous (GgAa) for both genes; we can test Carolina’s hypothesis against F2 data collected last fall. 4. From parents that are heterozygous for both green leaves and anthocyanin pigment, determine all the possible genotypes of offspring. (Brainstorm or use foil or two punnetts) Sum those that produce the same phenotype (green with anthocyanin; green, no anthocyanin; yellow, anthocyanin; yellow, no anthocyanin) and determine the predicted frequency of each. Multiply each predicted frequency by the number of plants the class grew and surveyed. Record in Table 3. 5. Actual data of each plant type from years of class results is found in Table 4. Use the chi-square formula to compare actual to expected for the 2012 data, for all years combined, and for each of the traits separately. Consult Table 2 to check the validity of the hypothesis. C. MORGANS FLIES : LINKED GENES Linkage is determined by crossing a hybrid organism with one that is recessive for both traits in a backcross. Recombination information regarding three genes on the same arm of the same chromosome is summarized in Table 4. Determine the relative location of each of gene. 6. Determine the expected frequencies of each phenotype from a cross of a heterozygote and a homozygous recessive if • the genes sort independently • the genes are on the same chromosome (linked) 7. Do Morgan’s data match either expected frequencies? Recombination of the parent pairs occurred but infrequently, indicating linked genes that occasionally are broken up by crossing over. The closer the genes are to one another on the chromosome, the more the traits show up together. The further apart, the more likely genetic recombination occurs. 8. Determine the percent recombination – phenotypes that do not match either parent – between each pair of traits. To find this value, use the following formula: recombined phenotypes ---------------------------- x 100 = percent recombination total offspring 9. Every 1% of recombination equals 1 Linkage Map Unit (LMU) between two traits. Use the three recombination percents to reflect the distances between the three genes on a chromosome. Sequence them on a number line to reflect their positions relative to one another. TABLE 1 : MENDELS’ MONOHYBRID CROSS RESULTS Parental traits Dominant Actual Dominant Actual Recessive Total plants Expected Dominant (.75) Expected recessive (.25) Chisquare result % likely the differences are due to chance Recessive Wrinkled White Constricted TABLE 4: MORGAN’S DATA dominant traits are in bold type. Black body & normal wing X Gray body & vestigial wing Offspring genotype Expected frequency -no linkage Actual result Expected result Purple eye & normal wing x Red eye & vestigial wing Black body & purple eye x Gray body & red eye BbVv Bbvv bbVv bbvv RrVv Rrvv rrVv rrvv BbRr Bbrr bbRr bbrr 0.25 0.25 0.25 .025 0.25 0.25 0.25 0.25 0.25 965 206 185 944 1339 151 154 1195 1159 0.25 0.25 0.25 67 78 1017 TABLE 1 : MENDELS’ MONOHYBRID CROSS RESULTS Parental traits Dominant Actual Dominant Actual Recessive Total plants Expected Dominant (.75) Expected recessive (.25) Chisquare result % likely the differences are due to chance Recessive Wrinkled White Constricted TABLE 4: MORGAN’S DATA dominant traits are in bold type. Black body & normal wing X Gray body & vestigial wing Offspring genotype Expected frequency -no linkage Actual result Expected result Purple eye & normal wing x Red eye & vestigial wing Black body & purple eye x Gray body & red eye BbVv Bbvv bbVv bbvv RrVv Rrvv rrVv rrvv BbRr Bbrr bbRr bbrr 0.25 0.25 0.25 .025 0.25 0.25 0.25 0.25 0.25 965 206 185 944 1339 151 154 1195 1159 0.25 0.25 0.25 67 78 1017 KEY: MORGANS FLIES : LINKED GENES Linkage is determined by crossing a hybrid organism with one that is recessive for both traits in a backcross. Recombination information regarding three genes on the same arm of the same chromosome is summarized in Table 2. Determine the relative location of each of gene. 1. Determine the expected frequencies of each phenotype from a cross of a heterozygote and a homozygous recessive if a) the genes sort independently (.25 each) b) the genes are adjacent on a chromosome (linked) (0.5 each) Table 2 dominant alleles are in bold type. Black body & normal wing X Gray body & vestigial wing Purple eye & normal wing x Red eye & vestigial wing Black body & purple eye x Gray body & red eye BbVv Bbvv bbVv bbvv RrVv Rrvv rrVv rrvv BbRr Bbrr bbRr bbrr 965 206 185 944 1339 151 154 1195 1159 67 78 1017 .09 .08 .41 etc .42 2. Do Morgan’s data match either expected frequencies? No. Recombination of the parent pairs occurred but infrequently, indicating linked genes that occasionally are broken up by crossing over. The closer the genes are to one another on the chromosome, the more the traits show up together. The further apart, the more likely genetic recombination occurs. 3. Determine the percent recombination – phenotypes that do not match either parent – between each pair of traits. To find this value, use the following formula: recombined phenotypes ---------------------------- x 100 = percent recombination total offspring ex: [(206 + 185)/2300] 100 = 16.9% recombination DISCUSSION: 1. In the garden pea, yellow cotyledon color is dominant to green, and inflated pod shape is dominant to the constricted form. Considering both of these traits jointly in self-fertilized dihybrids, the progeny appeared in the following numbers: 193 green, inflated 184 yellow constricted 556 yellow, inflated 61 green, constricted • Do these genes assort independently? Support your answer using Chi-square analysis. DISCUSSION: 1. Given parents AABbCc and AabbCc, how likely is the offspring Aabbcc? AABbCc? 2. Two grey cats have a litter of 8 kittens. 3 kittens are grey, 3 are white and 2 are black. • What genotypes might account for all these different colored kitties? Predict the genotype of the parent cats, and the probability of black, grey and white kittens in a litter of 8. • Does this example follow Mendel’s rules of dominance and recessiveness? Explain. • Use the chi-square formula to test your prediction with the data from the litter.



![Biology Chapter 3 Study Guide Heredity [12/10/2015]](http://s3.studylib.net/store/data/006638861_1-0d9e410b8030ad1b7ef4ddd4e479e8f1-300x300.png)