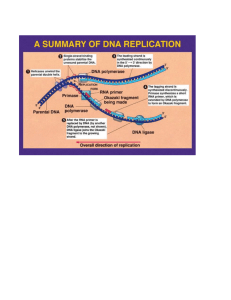
File - Chad M. Hodge
advertisement

Homework 3 – sequence alignments Chad Hodge – u0584663 BMI 6030 – Dr. Eilbeck 1. If you were looking for a mutation between your protein and a paralogous protein would you try a global or a local alignment first? (2 points) a. I think global would be the best one to try first. b. Explain your reasoning. (2 points) i. I would choose a global alignment because global alignments try to align proteins from the beginning to the end of the sequence. Since the mutations will be small, the global will show where the insertions or deletions or substitutions will pop out. Local alignment is meant for a smaller area of comparison, such as a motif, but global is meant for larger area matches, such as an entire sequence. 2. You search a 100-nucleotide long transcript against an EST database. The bit score of the best alignment was 37 bits. The effective database size was 100,000,000 nucleotides. The effective query size was 80 nucleotides. K = 0.059. a. Is the hit significant? (2 points) i. No, because the P value is greater than .05 significance. b. What is its E-value? (3 points) λ i. The Karlin-Altschul equation for this is normally shown as : E = Kmne- s however, since that equation calls for a raw score, and we have a bit score, I would suspect we should use this formula : E = m’n’2-s’ Solving this equation, we get this: E = 80*100,000,000*2-37 E = 0.05820766091 c. What is its P value? (3 points) i. The equation is this: P = 1-e-E Solving for this, we get: P = 1-e-0.05820766091 P = 0.05654599142 3. You want to look for distant homologs of your favorite Human protein in the Mouse Genome sequence. a. Which BLAST should you use (blastn, blastx, tblastn, tblastx, blastp)? (1 point) i. We are comparing protein (human) to nucleotide (mouse), across species, so we could use tblastn. If we converted our protein to a nucleotide, we could use tblastx to compare nucleotide to nucleotide, but this will introduce errors caused by splicing. b. Which BLOSUM Matrix(s) should you use? (3 points) i. BLOSUM62. I chose this because mice are a model organism, so are pretty similar to humans, so this could be a good starting point. c. Which BLOSUM Matrix would you use for searching the E. coli genome sequence with a human protein? (2 points) i. BLOSUM45 most likely, because humans and E. Coli are widely different. We could also look to BLOSUM62 if we think the gene is conserved between the 2. d. Why? (2 points) i. The lower the BLOSUM matrix number, the less identity match is used. This means that species that diverged long ago will have less in common with humans today, which E.coli would certainly fall under.