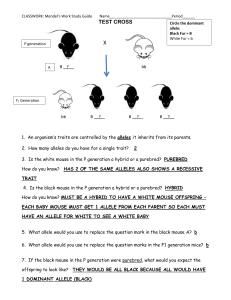
Methods
advertisement

Supplementary Methods Exome sequencing Exome libraries were prepared using the Agilent Sure Select All Exon 50Mb kit (Agilent Technologies, CA, USA) from isolated DNA. Each library was run in a single lane on a HiSeq2000 (Illumina Inc. CA, USA) using 100bp PE sequencing, at Canada’s Michael Smith Genome Sciences Centre. Exomes were aligned to hg19 using BWA-MEM (Li, 2013). Variants were called using the Genome Analysis Toolkit (1), and filtered for Mapping Quality ≥ 40, Quality by Depth ≥ 2.0, and Fisher Strand test ≥ 200. Functional annotations for predicted pathogenicity were run using snpEff (2). Mitochondrial Sequence Analysis In this analysis, the primary allele refers to the most common allele in the read counts, and the secondary allele refers to the second most common allele in the read counts. The reference allele refers to the allele of the rCRS reference sequence at a given position, while the alternate allele refers to the most common allele in the read counts that is not the reference allele. The total allele count refers to the sum of all alleles at a given position, excluding any ‘n’ calls. Mitochondrial reads were extracted from the whole exome sequencing data using MitoSeek (3), which was modified for analysis of both heteroplasmic and homoplasmic variants. An hg19 to rCRS conversion step was implemented to ensure that variant analysis would be performed relative to the rCRS reference sequence (NC_012920). Poor quality reads were filtered out on the criteria of Mapping Quality Score ≥ 20 and Base Quality Score ≥ 20. Positions with read depth ≤ 5 were considered unsuitable for variant analysis and excluded. A minimum secondary allele frequency of 10% and alternate allele count of 5 were set as the frequency cut-off values. When a potential heteroplasmy was called, the surrounding nucleotides were manually assessed with consideration to previously reported sequencing error trends (4,5) to determine whether variants were located in an ‘error hotspot’. Heteroplasmic variant calls that were outside of an error hotspot and that passed the 10% secondary allele cutoff as well as the 5-read alternate allele cutoff were taken as true heteroplasmies. Positions that did not meet the 10% secondary allele frequency cut-off were then evaluated as possible homoplasmic variants by comparing the primary allele with the reference allele at that position. Heteroplasmic frequencies of such variants were reported as (Alternate Allele Count)/(Total Allele Count). Amino acid differences were also reported for non-synonymous variants, accounting for the differences in genetic code between nuclear and mitochondrial DNA. HaploGrep (6,7) was used to predict the subjects’ haplogroup from the list of variants and to identify variants not accounted for by the assigned haplogroup. Global minor allele frequencies of reported variants were estimated using MITOMAP (8), which reported the frequency of each variant using 26850 mitochondrial GenBank sequences. Supplementary References 1. 2. 3. 4. 5. 6. 7. 8. McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. Sep 2010;20(9):1297-1303. Cingolani P, Platts A, Wang le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). Apr-Jun 2012;6(2):80-92. Guo Y, Li J, Li CI, Shyr Y, Samuels DC. MitoSeek: extracting mitochondria information and performing high-throughput mitochondria sequencing analysis. Bioinformatics. May 1 2013;29(9):1210-1211. Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. Sep 2008;36(16):e105. Li M, Stoneking M. A new approach for detecting low-level mutations in nextgeneration sequence data. Genome Biol. 2012;13(5):R34. Kloss-Brandstatter A, Pacher D, Schonherr S, et al. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum Mutat. Jan 2011;32(1):25-32. van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. Feb 2009;30(2):E386-394. Ruiz-Pesini E, Lott MT, Procaccio V, et al. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. Jan 2007;35(Database issue):D823-828.