june_2014_newsletter - BEA core facility
advertisement

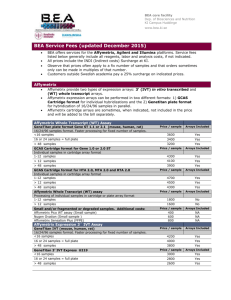
BEA is now a SciLifeLab Regional Facility New Protocols from Affymetrix BEA, the Core Facility for Bioinformatics and Expression Analysis (www.bea.ki.se/), a SciLifeLab regional facility of national interest, supporting research for genomic analysis with microarrays and sequencing methods. BEA has during 2014 introduced the Sensation Plus FFPE Amplification and WT Labeling protocols. This complete solution for FFPE extracted RNA allows amplification and labeling from as little as 20 ng of total RNA. For other problematic or small samples we continue to offer the Nugen Ovation Pico amplifications for WT and IVT arrays which allows expression profiling starting with as little as 500 pg total RNA. New Arrays from Affymetrix Traditional microarray analysis methods offer fast, economical and comprehensible solutions. New products releases from Affymetrix include the new transcriptome arrays for mice (MTA available Q3 2014) and rats (RTA available Q4 2014). The already available Human Trancriptome array (HTA) maximizes the amount of unique and valuable information with a high-resolution array design which contains >6.0 million probes covering coding transcripts and non-coding transcripts covering exons for coding transcripts and exon-exon splice junctions and non-coding transcripts. The most economical and high throughput solutions are the updated whole transcript Human Gene 2.1 ST Array plate (3400 Skr/sample) that include probes for many non-coding RNA transcripts and the miRNA 4.1 Array plate (2000 Skr/sample) array. Sequencing at BEA If you are interested in performing a sequencing project at BEA we are offering complete genomic solutions with the Illumina Miseq and Hiseq platforms in Huddinge. The service can be adapted for your needs and we provide different library construction and sequencing methods. Please inquire for details about different library protocols and sequencing pricing. Illumina Sequence platform Summer Vacation BEA will be partly closed week 30-31 (July 21 - August 1). The core facility staff will be reduced during the rest of the summer, and we strongly recommend visitors to contact the core facility before visiting. New Software from Affymetrix Affymetrix has also released a new version of the Transcriptome Analysis Software, TAC 2.0, allowing analysis of Interaction Network between miRNA and mRNA arrays and analysis of splicing event estimate to classify splicing analysis result from the Affymetrix transcriptome and exon arrays. We Wish You All a Very Nice Summer! Bioinformatics and Expression Analysis core facility (BEA) Fredrik Fagerström-Billai Marika Rönnholm Malin Liljebäck Susann Fält Patrick Müller David Brodin Group Leader Research Engineer Research Engineer Research Engineer Research Engineer Bioinformatician fredrik.fagerstrom-billai@ki.se marika.ronnholm@ki.se malin.liljeback@ki.se susann.falt@ki.se patrick.muller@ki.se david.brodin@ki.se Visiting Address Department of Biosciences and Nutrition at NOVUM, Karolinska Institutet, SE-141 57 Huddinge, SWEDEN Homepage: www.bea.ki.se