jane12210-sup-0001
advertisement

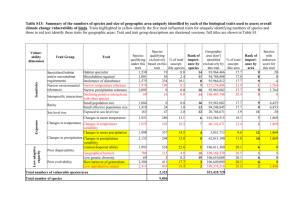
Details of individual housing and feeding Food was placed in the open area of the tank, furthest from the shelter tube. Individuals were fed daily a varied diet of yabby feed (see above) and additionally frozen peas and bloodworms on alternate days (Jerry et al. 2002). In contrast to many fish species, yabbies will eat food that rests on bottom and consume food that has been in the tank for many hours. Ensuring excess food was present was critical for any differences in growth to reflect their intrinsic growth capacity (see Discussion). Each day between 1300h and 1500h, uneaten food and faeces were removed using a fine mesh net, and fresh food added. Full water changes were carried out once weekly, and half water changes mid-week. Nonetheless, we monitored water temperature, dissolved oxygen and ammonia levels every third day in 12 aquaria that housed individuals known to be consuming the most and least amounts of food (fastest and slowest growers). Oxygen never fell below 7mg/L and ammonia never rose above 0.1ppm during the experiment. Because temperature varied 1.5°C across the length of the lab bench, we moved tanks semi-randomly each week to avoid systematic bias with respect to growth (temperature effects on behaviour, by contrast, could be controlled for in our models, see below). Details of statistical analyses The mixed models used here are often termed random regression, generating predictions for individual-specific deviations from the mean-level intercept and slope(s) (i.e. the socalled ‘BLUPs’, best linear unbiased predictors. We specified an ‘unstructured’ variance- covariance matrix, meaning that ‘covariance’ parameters were fit to describe correlations between individual intercept and slope values (covis) in addition to the intercept (vari) and slope (vars) variance parameters. We tested whether families differed with respect to their intercepts and slopes of behavior across days, or time of day, by specifying these predictors as being random with respect to family identity, and nesting individuals within family (see Singer 1998 for a worked example). We arrived at the final best model by sequentially culling factors one at a time that were not significant (P > 0.1), starting with those parameters with the largest p-value (Crawley 2005). The fixed effect of sex was evaluated first, then random effects associated with slope parameters. All analyses were implemented using SAS ‘Proc Mixed’, using ReML, where fixed factors are evaluated using F-tests, random effects tested using z-tests (which are conservative relative to a one-tailed likelihood-ratio test), the Kenward-Roger method to calculate denominator degrees of freedom for the fixed effects (which can generate noninteger df values), and a type III sums of squares approach for fixed effects (Littell et al. 2006). Other statistics packages often report χ2 values (df =1) for variance components, but these are often simply the z-value squared (Singer & Willett 2003). Boldness and voracity were ln-transformed to normalise data for analysis. Because at least one random slope effect was significant in all analyses, we could not calculate repeatability values in the ‘standard’ way, because the rank-order of individuals, their ‘personality’, changed across contexts (over days, day versus night, temperature). Therefore, we followed the model of summing variance as presented by (Singer & Willett 2003), and included additional terms to accommodate the additional variance terms (and associated covariances) for up to two random slope effects, to generate the following equation to calculate repeatability specific to each random slope effect: R = (variance across individuals) / (variance across + variance within individuals) = (varint + 2*covis*X1 + vars*X12 + 2*covis*X2 + 2*covss* X1*X2 + vars*X22 ) / (varint + 2*covis*X1 + vars*X12 + 2*covis*X2 + 2*covss* X1*X2 + vars*X22 + varresidual) where varint is the random intercept variance, vars is random slope variance, covis is a covariance between intercepts and slopes, covss is the covariance between slopes, and varresidual is the residual (within-individual) variance; X1 and X2 represent two different variables, one for each random slope effect. Therefore, for example, the term “vars*X22” represents the variance of the second random slope effect, and “2*covss* X1*X2” represents the covariance between individual slopes related to the random slope effects for variables X1 and X2. To estimate a growth rate and BT value for each individual for subsequent analyses, we extracted model parameters, and individual predicted deviations (BLUPs) from the mean-level parameters, into a spreadsheet. There, we calculated predicted values for each yabby that accounted for the effect of any significant covariates. In the case of boldness we calculated BT values for day and night because individuals differed in their diurnal responses (random slope effect). There was an effect of date on boldness in the first set of boldness trials (April to June), but not the second (July), and so we fixed date equal to the mid-point of each set of observations when calculating BTs. Temperature had effects on boldness and voracity and so we set temperature equal to 19C in the model when deriving BT predictions, as this was the mean temperature in May through June observations (n = 1979). To ensure the model was fitting the data correctly and assumptions of the models were not violated, we plotted observed values in relation to the model-predicted values, both in relation to time of day, and in relation to observation day (Singer & Willett 2003). Because we were unable to determine the sex for 4 individuals, and sex was a significant predictor in our mixed models, our effective sample size was n = 86. Results of multivariate mixed model using mass (random effects portion only). Estimated G Matrix Row 1 2 3 4 Effect trait id trait trait trait trait bold-day 1 bold-night1 voracity 1 mass 1 Col1 Col2 Col3 Col4 0.3905 0.3726 -0.2687 0.2747 0.4699 -0.3293 0.3566 0.3793 -0.2252 0.3688 Estimated G Correlation Matrix Row 1 2 3 4 Effect trait id Col1 Col2 Col3 Col4 trait trait trait trait bold-day 1 bold-night1 voracity 1 mass 1 1.0000 0.8698 -0.6981 0.7238 1.0000 -0.7798 0.8567 1.0000 -0.6020 1.0000 Covariance Parameter Estimates Cov Parm Subject UN(1,1) UN(2,1) UN(2,2) UN(3,1) UN(3,2) UN(3,3) UN(4,1) UN(4,2) UN(4,3) UN(4,4) Residual Residual Residual Residual id id id id id id id id id id id id id id Group Estimate Standard Error Z Value Pr Z trait trait trait trait 0.3905 0.3726 0.4699 -0.2687 -0.3293 0.3793 0.2747 0.3566 -0.2252 0.3688 bold-day 0.5369 bold-night 0.5105 voracity 0.6012 mass 0.2359 0.06665 0.06777 0.08861 0.05653 0.06622 0.07026 0.05655 0.06754 0.05499 0.06956 0.02365 0.03898 0.03468 0.02551 5.86 5.50 5.30 -4.75 -4.97 5.40 4.86 5.28 -4.09 5.30 22.70 13.10 17.33 9.25 <.0001 <.0001 <.0001 <.0001 <.0001 <.0001 <.0001 <.0001 <.0001 <.0001 <.0001 <.0001 <.0001 <.0001 Correlation matrix result from multivariate mixed model when using growth rate (rather than mass). Estimated G Correlation Matrix using growth rate Row 1 2 3 4 Effect trait id trait trait trait trait bold-day bold-night growth voracity 1 1 1 1 Col1 Col2 Col3 Col4 1.0000 0.8698 0.6587 -0.6981 0.8698 1.0000 0.8344 -0.7798 0.6587 0.8344 1.0000 -0.6373 -0.6981 -0.7798 -0.6373 1.0000