Computational Methods
advertisement

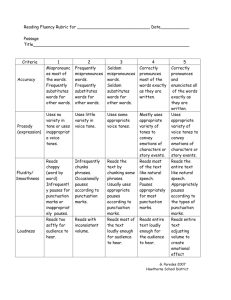
I. Lariat Discovery Illumina Dataset. To identify lariat branchpoints, we analyzed the Illumina Human Body Map 2.0 total RNA deep sequencing library. Reads consist of RNA samples derived from 16 human tissues. Most reads are 100 bp in length and all linker sequences were removed prior to analysis. Hg19 Annotated. We discovered branch points by searching for reads with non-canonical arrangements of intronic sequences. Reads that aligned to the hg19 genome with 3 or less mismatches were eliminated. Each remaining read was split into all possible head and tail segments, in which all heads and tails were at least 15 nt long. We mapped all head and tail segments to the hg19 genome using bowtie. Head and tail segments were required to map to only one place in the genome and without any errors. Reads with segments that mapped in the expected order or did not map intronically were filtered from the dataset. The remaining inverted reads were mapped to splice sites. In cases where the read tail begins at the first nucleotide of the intron and the read head mapped within 500 nt upstream of a 3’ss, we determined that the read spans the lariat 2’-5’ lariat linkage. In cases where there is alignment ambiguity, we allow up to two mutations and assume the alignment in which the tail maps to the first nucleotide of the intron. The last nucleotide of the read head was determined to be the branchpoint. 2066 lariat reads were discovered through this screen (lariat_0 – lariat_2065 in BED track). Reads that suggested branchpoints supported by spliced EST evidence were also reported. Nine lariat reads were discovered through this screen (lariat_2110 – lariat_2118 in BED track). Unannotated, but with Illumina transcript support. To discover lariats forming in transcripts that are unannotated in the hg19 assembly, we built a library of potential spliced products inferred from the inverted reads. For each inverted read that did not map to an annotated 5’ss, we constructed a potential upstream exon by taking an 85 nucleotide window immediately upstream of the read tail. We created an array of 200 potential downstream exons by taking 85 nucleotide windows at a distance of 1 to 200 nucleotides from the end of the read head. These windows were artificially spliced together to create a set of potential spliced products. We aligned the Illumina reads against these spliced products using bowtie, requiring that the read contained at least 15 nucleotides on either side of the splice junction and did not have any mismatches. In cases where a splice product was found and the implied intron contained a 5’ GT and 3’ AG sequence, the inverted read was determined to be a true lariat forming in an unannotated transcript. Forty-four additional lariat reads were discovered through this screen (lariat_2066 – lariat_2109 in BED track). Lariats forming deep within introns. The remaining out-of-order reads with intronic heads and tails, but without annotated or Illumina transcript support, were studied. From these reads, we filtered a high confidence set of lariats by requiring that both the heads and tails were never annotated as exons (alternative events), the beginning of the tail had a patser score of at least 6.0 against a 5’ss position specific weight matrix, and the read had a mutation at the branchpoint. The 5’ss position specific weight matrix was created by inputting all hg19 annotated 5’ss sequences into the patser program. From within this high confidence set of internal lariats, the mutational profile was similar to the mutational profile of the bona-fide lariats (mostly A-> T mutations). We counted the number of splicing events that used an annotated 5’ss without a 3’ss, an annotated 3’ss without a 5’ss, or an event deep within an intron (using no annotated splice sites). We also counted the number of bona-fide lariats that passed the patser score, mutational, and intronic filters, and used that fraction to extrapolate how many true lariats without transcript support we expect exist in our data. As a control, in-order reads that were most likely caused from template switching were passed through these same filters. There is a 3.3 fold increase of out-of-order reads that pass these filters compared to in-order reads. Also, the mutational profile of the in-order reads was different than bona-fide lariats. This suggests that these internal lariats are truly forming. II. Analysis Branch Point Characterization. The branch point distance was measured as the distance between the last nucleotide of the read head to the first downstream annotated 3’ splice site. The mutational profile of reads that suggested a branch point at the last nucleotide of the intron (implying a circular intron) were compared to the mutational profile of all other reads with ‘G’ nucleotide branchpoint more distal from the 3’ss. A chi-squared test was used to show that these mutational profiles were significantly different. mRNA exon junction data was studied using tophat. We created a junction file consisting of all possible constitutive and exon skipping events within each annotated transcript. We aligned the Illumina reads using the hg19 genome and this junction file to determine how many reads span each exon/exon junction. We calculated overall rates of alternative splicing and intersected this data with our lariat branch points. In cases where the lariat formed over a skipped exon or immediately upstream of a skipped exon, the branchpoint distance was measured to the first downstream exon. In cases where the lariat formed near an alternative 3’ss, the branchpoint distance was measured to the most upstream 3’ss. For all three classes of alternative events, p values were determined by randomly sampling the entire dataset 1000 times and counting how many times the average branchpoint distance was at least as extreme as the branchpoint distance in the alternative event. Splice Site Recognition Analysis. The number of ‘AG’ dinucleotides between the branchpoint and the 3’ss ‘AG’ were counted for the 2066 hg19 annotated lariats. 2066 introns were selected at random, and simulated branchpoints were selected by randomly distributing the branchpoint distance distribution that was observed in real lariats. The number of ‘AG’ dinucleotides between these simulated branchpoints and the 3’ss ‘AG’ were counted. This process was repeated 1000 times to generate a p-value. The AG selection decision tree analysis was performed using C5.0 decision tree software. Lariat introns with exactly one branchsite and one used 3’ss were considered in this analysis. The used AG, immediately upstream AG (if existant), and downstream AG were included in the dataset. First, the data was organized into a decision tree using classifiers from the literature, including distance to branchsite, distance to upstream and downstream AGs, the nucleotide upstream of the AG, and presence of secondary structure (Gibbs free energy determined using RNAfold). In this initial run, the only informative classifiers were the presence or lack of an upstream AG, the distance between the AG and the branchsite, and the distance to upstream and downstream AGs. Next, a range of constraints for each of the distance classifiers were applied to the dataset, and C5.0 was run on each of combination of classifier constraints. The classifier sets with error rates lower than the literature classifier sets were run again on the dataset, this time using half of the dataset as training data, and the other half as testing. The highest predictive scoring classifier sets were subjected to 10-fold cross validation trials. These cross-validation trials were completed 1000 times. The classifier set with the highest average predictive accuracy was used to create the decision tree. RNA-protein Interactions. Published CLIP data for FOX2 (Yeo et. al, 2009), PTB (Xue et. al,2009), and hnRNP C (König et. al, 2010) were mapped around our branchpoint coordinates. The FOX2 and PTB data was smoothed by using the center CLIP coordinate and adding 15 nucleotides to either side. The raw hnrnpC CLIP reads were aligned using bowtie and the last nucleotide was used as the binding point (as described in study). We smoothed the hnrnpC data by adding 15 nucleotides to either side of the binding point. Lariat Recovery. The number of reads spanning each annotated exon/exon junction were determined using tophat. Lariats and exon/exon junctions were both binned by intron size. The recovery rate of a lariat read was calculated by counting the number of detected lariat reads and dividing it by the number of detected exon/exon junction reads within each intron bin. The error bars were calculated by resampling the lariat read data 1000 times and using the 95% confidence interval.