Protocol for inactivation of the genes in Salmonella Typhimurium
advertisement
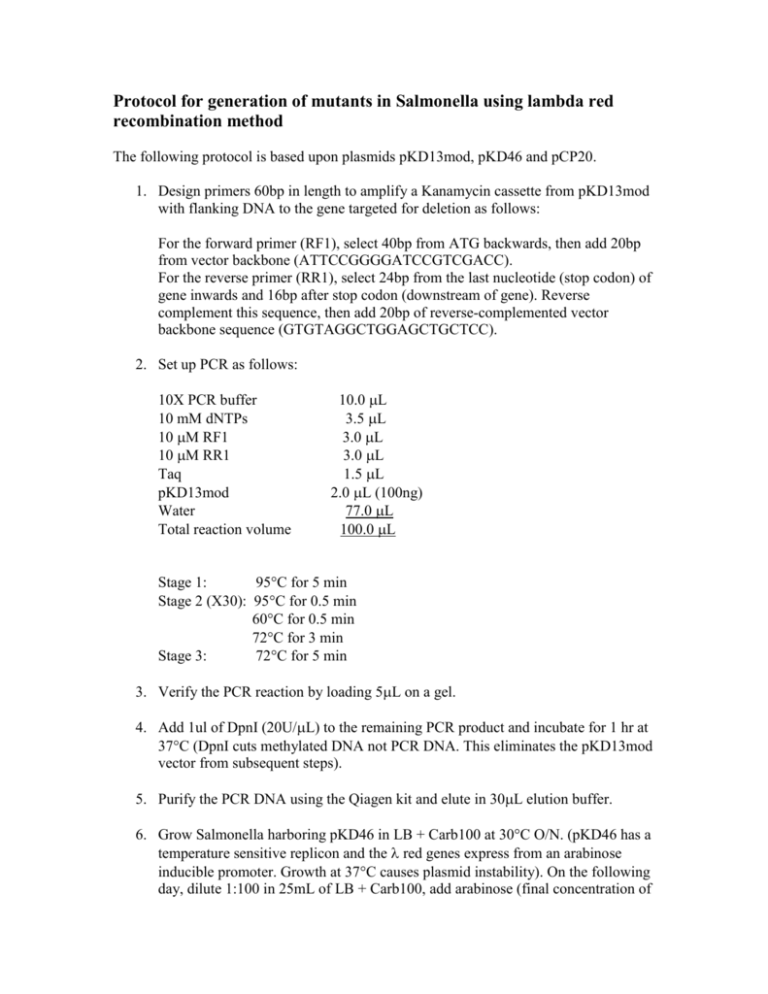
Protocol for generation of mutants in Salmonella using lambda red recombination method The following protocol is based upon plasmids pKD13mod, pKD46 and pCP20. 1. Design primers 60bp in length to amplify a Kanamycin cassette from pKD13mod with flanking DNA to the gene targeted for deletion as follows: For the forward primer (RF1), select 40bp from ATG backwards, then add 20bp from vector backbone (ATTCCGGGGATCCGTCGACC). For the reverse primer (RR1), select 24bp from the last nucleotide (stop codon) of gene inwards and 16bp after stop codon (downstream of gene). Reverse complement this sequence, then add 20bp of reverse-complemented vector backbone sequence (GTGTAGGCTGGAGCTGCTCC). 2. Set up PCR as follows: 10X PCR buffer 10 mM dNTPs 10 M RF1 10 M RR1 Taq pKD13mod Water Total reaction volume 10.0 L 3.5 L 3.0 L 3.0 L 1.5 L 2.0 L (100ng) 77.0 L 100.0 L Stage 1: 95C for 5 min Stage 2 (X30): 95C for 0.5 min 60C for 0.5 min 72C for 3 min Stage 3: 72C for 5 min 3. Verify the PCR reaction by loading 5L on a gel. 4. Add 1ul of DpnI (20U/L) to the remaining PCR product and incubate for 1 hr at 37C (DpnI cuts methylated DNA not PCR DNA. This eliminates the pKD13mod vector from subsequent steps). 5. Purify the PCR DNA using the Qiagen kit and elute in 30L elution buffer. 6. Grow Salmonella harboring pKD46 in LB + Carb100 at 30C O/N. (pKD46 has a temperature sensitive replicon and the red genes express from an arabinose inducible promoter. Growth at 37C causes plasmid instability). On the following day, dilute 1:100 in 25mL of LB + Carb100, add arabinose (final concentration of 0.1M) and grow at 30C for 4 hrs. Wash cells 4 times in ice-cold water and resuspend in residual liquid. Transfer 80L to ice-cold electroporation cuvettes and add about 800ng of corresponding PCR product. Leave on ice for 5 min. Electroporate and add 350L of SOC. Transfer to tube and shake for 1 hour at 30C. Then centrifuge cells, discard supernatant and resuspend pellet to a final volume of 100L. Spread cells on LB-Kan60 plates and incubate O/N at 37C. 7. Design primers to confirm that Kan gene has replaced the gene of interest: For the forward primer (DF1), select 20 or 21bp upstream of the gene that’s been deleted (not too close to gene sequence). For the reverse primer, use K1 that binds to the Kan marker. (K1: CAGTCATAGCCGAATAGCCT is a reverse primer that anneals to the start of the Kan marker) 8. Next day, pick at least 2 colonies from the LB-Kan60 plate and resuspend in 100L of water. Do colony PCR as follows: 10X PCR buffer 10 mM dNTPs 10 M DF1 10 M K1 Taq Resuspended colony Water Total reaction volume 10.0 L 3.5 L 3.0 L 3.0 L 1.5 L 1.0 L 78.0 L 100.0 L Stage 1: 95C for 5 min Stage 2 (X30): 95C for 0.5 min 50C for 0.5 min 72C for 3 min Stage 3: 72C for 5 min 9. Take 2L of the resuspended colony and spot it on LB-Kan60 plate and incubate plate at 43C O/N to ensure loss of pKD46. 10. Run PCR on a gel (product should be ~500bp). 11. Restreak PCR-positive colony from step 9 on LB-Kan60 plate 2 times over 2 days. On the final day, streak on both LB-Kan60 and LB-Carb100 plate. Repeat till no growth is seen on LB-Carb100 plate. Make freezer stock. 12. Inoculate colony from previous step in LB + Kan60 and grow at 37C O/N. On the following day, take 5mL of LB + Kan60, then add 0. 05mL of the O/N culture and incubate at 37C for 3 hours. Wash cells 4 times in ice-cold water and resuspend in residual liquid. Transfer 80L to ice-cold electroporation cuvette and add 100ng of pCP20. Leave on ice for 5 mins. Electroporate and add 350L of SOC. Transfer to tube and shake for 1 hour at 30C. Then centrifuge cells, discard supernatant and resuspend pellet to a final volume of 100L. Spread cells on LB-Carb100 plate and incubate for 2 days at 30C. 13. To confirm deletion of gene, do colony PCR using DF1 and DR1 (DR1 is reverse primer obtained by selecting 20 or 21 bp downstream of the gene that’s been deleted (not too close to gene sequence) and reverse complemented). Pick at least 2 colonies from the previous step and resuspend in 100L of water. Do colony PCR as follows: 10X PCR buffer 10 mM dNTPs 10 M DF1 10 M DR1 Taq Resuspended colony Water Total reaction volume 10.0 L 3.5 L 3.0 L 3.0 L 1.5 L 1.0 L 78.0 L 100.0 L Stage 1: 95C for 5 min Stage 2 (X30): 95C for 0.5 min 50C for 0.5 min 72C for 3 min Stage 3: 72C for 5 min 14. Take 2L of the resuspended colony and spot it on LB plate and incubate at 43C O/N. Run PCR from previous step on a gel (PCR product should be ~500bp). If product of the right size is seen, purify the product using Qiagen kit then confirm deletion by DNA sequencing. 15. Restreak colonies from previous step on LB plate, 3 times over 3 days. On the final day, streak on both LB and LB-Carb100 plates. Repeat till no growth is seen on LB-Carb100. Make freezer stock.







