Chi-square with R
advertisement

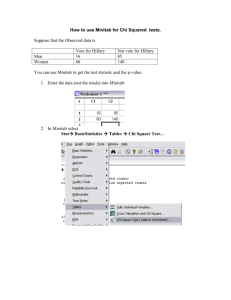
From: http://statland.org/R/Rchisq.htm 1. Chi-square with R R can do all the usual chi-square tests, with either raw data or tables of counts. We start with the heart attack data and the Goodness-of-Fit test. > names(heartatk) [1] "Patient" "DIAGNOSIS" "SEX" [7] "LOS" "AGE" "DRG" "DIED" "CHARGES" > attach(heartatk) > table(SEX) SEX F M 5065 7779 > chisq.test(table(SEX)) Chi-squared test for given probabilities data: table(SEX) X-squared = 573.4815, df = 1, p-value < 2.2e-16 The command chisq.test(table(SEX)) does a chi-square goodness of fit test on the table for the SEX variable. The default is to test for equal expected counts in every cell. That is hardly the case here. 2.2e-16 means 2.2X10-16 = 0.00000000000000022, which is small. If you do not want equal proportions, you need to give a set of proportions for each cell. For example, genetic theory predicts that certain fruit flies will fall into four categories in proportions 9:3:3:1. Data showed counts of 59, 20, 11 and 10. > chisq.test(c(59,20,11,10),p=c(9/16,3/16,3/16,1/16)) Chi-squared test for given probabilities data: c(59, 20, 11, 10) X-squared = 5.6711, df = 3, p-value = 0.1288 We would not reject the theoretical hypothesis with these data. You can make a cross-tabulation of two categorical variables with table and do a test of independence or homogeneity with chisq.test. (We return to the heart attack data.) > table(SEX,DIED) DIED SEX 0 1 F 4298 767 M 7136 643 > chisq.test(table(SEX,DIED)) Pearson's Chi-squared test with Yates' continuity correction data: table(SEX, DIED) X-squared = 147.7612, df = 1, p-value < 2.2e-16 Page 1 of 4 We have seen this p-value before! It is probably the smallest non-zero number R can handle and hence not very accurate. However, p is definitely small! Hence we reject the hypothesis that the mortality rate is the same for men and women. Looking at the data, it is higher for men. With 12,844 observations, getting the table is a lot more work than computing chi-square, and it is best to let the computer do it. If you have an existing table, R will analyze it -- but not without putting up a fight. You need to enter the table one row at a time and use rbind to combine the rows into a table. Here is some data on hepatitis C incidence (yes,no) and tattoo "status": from a parlor, elsewhere, and no tattoo. > hep=rbind(c(17,35),c(8,53),c(22,491)) > hep [,1] [,2] [1,] 17 35 [2,] 8 53 [3,] 22 491 > chisq.test(hep) Pearson's Chi-squared test data: hep X-squared = 57.9122, df = 2, p-value = 2.658e-13 Warning message: Chi-squared approximation may be incorrect in: chisq.test(hep) The warning message probably refers to the small expected count the cell where the 8 appears. You could overcome that by pooling the two tattoo sources together. Either way, getting tattoos seems to greatly increase the risk of hepatitis C. http://statland.org/R/ 2. Reading Tables into R In school, we work mainly with tiny datasets we can type into our technology if all else fails. In practice, we generally start with raw data in a computer file. Often, getting the data into a form our technology can work with is a major undertaking. We will go through that once right now. R can read data from a text file. The text file has to be in the form of a table with columns representing variables. All columns must be the same length. Missing data must be signified by "NA". Optionally, the first row of the file may contain names for the variables. You can access such a file named heartatk4R.txt. Download and save this file in the directory where the R program lives. The file looks like this. Patient 1 2 3 4 5 6 7 8 9 10 11 12 DIAGNOSIS 41041 F 41041 F 41091 F 41081 F 41091 M 41091 M 41091 F 41091 F 41041 M 41041 F 41041 F 41091 M SEX 122 122 122 122 122 121 121 121 121 123 121 121 DRG 0 0 0 0 0 0 0 0 0 1 0 0 DIED CHARGES 4752 0010 3941 0006 3657 0005 1481 0002 1681 0001 6378.6400 10958.520 16583.930 4015.3300 1989.4400 7471.6300 3930.6300 LOS 079 034 076 080 055 0009 0015 0015 0002 0001 0006 0005 AGE 084 084 070 076 065 052 072 Page 2 of 4 13 14 15 16 17 41091 41091 41041 41041 41041 F F M M M 122 122 122 122 121 0 0 0 0 0 ¥ 0009 4433.9300 3318.2100 4863.8300 5000.6400 083 0004 0002 0005 0003 061 053 077 053 Above are only the first 17 cases (of 12,844). To use this in R you must define a variable to be equal to the contents of this file. > heartatk = read.table("heartatk4R.txt",header=TRUE) The argument header=TRUE tells R that the first row of the file should be interpreted as variable names. (These should not include spaces.) You can now get a table of contents for what you have created in R with > objects() This should return heartatk along with any other variables you may have created. You will not see on this list any of the variables that are inside of heartatk because they are hiding. To see them, type > names(heartatk) [1] "Patient" "DIAGNOSIS" "SEX" [7] "LOS" "AGE" "DRG" "DIED" "CHARGES" To bring them out of hiding, you must "attach" them to your R workspace. (This avoids conflicts if several tables include variables with the same name. Attach just one at a time.) > attach(heartatk) These data came from an ActivStats CD which provided this background information: Heart Attack Patients This set of data is all of the hospital discharges in New York State with an admitting diagnosis of an Acute Myocardial Infarction (AMI), also called a heart attack, who did not have surgery, in the year 1993. There are 12,844 cases. AGE gives age in years SEX is coded M for males F for females DIAGNOSIS is in the form of an International Classification of Diseases, 9th Edition, Clinical Modification code. These tell which part of the heart was affected. DRG is the Diagnosis Related Group. It groups together patients with similar management. In this data set there are just three different drgs. 121 for AMIs with cardiovascular complications who did not die. 122 for AMIs without cardiovascular complications who did not die. 123 for AMIs where the patient died. LOS gives the hospital length of stay in days. DIED has a 1 for patients who died in hospital and a 0 otherwise. CHARGES gives the total hospital charges in dollars. Data provided by Health Process Management of Doylestown, PA. After you attach the data table you can work with the internal variables providing you remember that R is casesensitive. > table(sex) Error in table(sex) : object "sex" not found > table(SEX) SEX F M Page 3 of 4 5065 7779 © 2006 Robert W. Hayden. Data Desk is a registered trademark of Data Description. Page 4 of 4