19 QTL analysis.korME.Cor2ME
advertisement

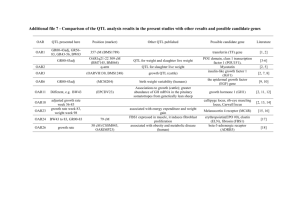
QTL Mapping QTL (Quantitative Trait Loci) analysis is the correlation of data from quantitative plant traits, as analysed in plant phenomics, with all marker-based or direct genetic data available for the plant. The greater this correlation is, the higher the probability that a certain gene contributes directly to a specific trait. The core aspect of the QTL method is its openness of experimental design towards other questions, even in cases where the researcher does not expect a result. This characteristic makes the technology interesting for the discovery of new aspects, especially as both quantitative gene data and phenotyping data have gained enormously in quality and quantity over the last years. The LemnaTec scanalyzer plant phenotyping concept fits perfectly into this research approach. QTL analysis and LemnaTec scanalyzer plant phenomics 1. LemnaTec image-based plant phenotyping offers a large amount of quantitative data for each individual plant. Such data can be put into QTL correlation processes at quite different levels of data transformation – from almost raw data (leaf colour or areas) to transformed data (growth rates) and complex, derived traits (e.g. drought resistance, water or nutrient efficiencies etc.). 2. All data is provided at a high frequency, which also allows using data from up- and down-regulated genes in an effort to understand the complex controls of plant development over time. 3. LemnaTec plant phenotyping raw data is acquired with a minimum of underlying research hypotheses, in order to be open to the unexpected. Thus, it provides the best chances for unanticipated findings, even when correlating complex phenomic traits to gene loci in a quantitative way. 4. As all raw data is stored, first hints of any correlation can be validated by recalling retrospective use from earlier experiments or by reanalysing the images more specifically. 5. Especially for extremely complex and multidimensional data like those resulting from hyperspectral imaging, the QTL approach is an excellent option to discriminate between relevant information and background noise. BU High-throughput phenotyping for QTL mapping under highly controlled plant development and imaging conditions provides the best basis for extracting a maximum of information from test objects. This way, reproducible, comprehensive datasets are generated that are open enough to let the plants themselves answer the question of relationship between their phenotypic traits and genes.