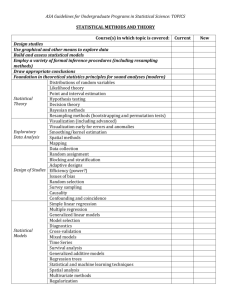
text01
advertisement

Generalized linear models Generalized linear models (GLMs) are mathematical extensions of linear models which are capable to handle non–linear relationships and various statistical distributions [McCullagh and Nelder, 1989]. In this study, GLMs were based on first and second order polynomials and logit –link function. In addition, to calibrate GLMs we utilized an automatic stepwise procedure based on the Akaike’s information criteria (AIC). Generalized additive models Generalized additive models (GAMs) are semi-parametric extensions of GLMs [Hastie and Tibshirani, 1990]. To relate the occurrence of ESPs to environmental predictors, the maximum smoothing function was set to 4 which were subsequently optimized by the model fitting function [Wood, 2011]. Generalized boosting method Generalized boosting method (GBM) is a sequential ensemble modeling method that combines a large number of iteratively fitted classification trees to a single model with improved prediction accuracy [Elith et al., 2008]. GBMs automatically incorporate interactions between predictors and are capable of modeling highly complex non–linear systems. GBMs (with Bernoulli–distribution assumption) were fitted setting the interaction depth to six, bagging fraction to 0.75 and maximum number of trees to 3000. Classification tree analysis Classification tree analysis (CTA) generates a binary tree through binary recursive partitioning i.e. the nodes are split based on true/false answers concerning the values of predictors [Venables and Ripley, 2002]. CTA builds an overgrown tree based on the node purity -criterion that is later pruned back via tenfold cross-validation to avoid over-fitting [Breiman et al., 1984]. Artificial neural network Artificial neural network (ANN) is a machine-learning method that replicates the functioning of the human brain. This highly flexible method builds accurate models for prediction when the functional form of the underlying equations is unknown [Venables and Ripley, 2002]. ANNs were fitted by setting the maximum number of iterations to 200 (Biomod 2 default). To prevent the ANNs to over fit, four–fold cross–validation was implemented to stop the training of the networks. Random forest The random forest (RF) generates a large ensemble of classification trees forming a “forest”. Each tree is subsequently built by randomly selecting a training dataset from the observations (i.e. bootstrap sample with replacement; n=1200 in this study). In addition, four explanatory variables in each tree were randomly selected for calculating the best split on these predictors in the training set [Breiman, 2001]. This procedure is iterated over all trees in the ensemble and the RF algorithm detects the classification appearing most frequently in the model selection process (i.e. the random forest prediction). In this study, we set the maximum number of trees to 500. Multivariate adaptive regression splines Multivariate adaptive regression splines (MARS) merge standard linear regression, mathematical spline construction and binary recursive partitioning in order to produce a local model in which relationships between response and explanatory variables are either linear or non-linear [Friedman, 1991]. In this study, we set maximum interaction degree to two, forward stepwise stopping threshold to its default value (0.001) with pruning of the model allowed. Maximum entropy Maximum entropy (MAXENT) is a flexible statistical modeling approach where a target probability distribution is estimated by finding the probability distribution of maximum entropy (i.e. that is most dispersed or closest to uniform) [Phillips et al., 2006]. It has many advantages over some other machine learning techniques e.g. the MAXENT probability distribution has a mathematical definition and is not sensitive to over-fitting [Phillips et al., 2004]. In our study, we set the maximum number of model iterations to 200. Flexible discriminant analysis Flexible discriminant analysis (FDA) is a non–parametric equivalent of the linear discriminant analysis (LDA) [Hastie et al., 1994]. FDA is a multi–group nonlinear classification technique that replaces the linear regression by any nonparametric method [Hastie et al., 1995]. The FDAs were optimally scaled by using MARS (default option in Biomod2). Surface range envelope Surface range envelope (SRE) is a range based technique where the algorithm identifies locations where all the predictors fall within the extreme values determined for a set of observations (97.5% in this study) [Carpenter et al., 1993]. SRE defines the suitable environmental conditions for target ESPs as a rectangular envelope in Euclidean space. Literature cited: Breiman, L., Friedman, F., Olshen, F., and C. Stone (1984), Classification and Regression Trees. Wadsworth Statistics/Probability Series. Wadsworth, Belmont, CA, USA, 358pp. Breiman, L. (2001), Random forests. Machine Learning 45(1), 5–32, doi: 10.1023/A:1010933404324 Carpenter, G., Gillison, A. N., and J. Winter (1993), DOMAIN: a flexible modelling procedure for mapping potential distributions of plants and animals. Biodiversity and Conservation 2(6), 667– 680, doi: 10.1007/BF00051966 Elith, J., Leathwick, J. R., and T. Hastie (2008), A working guide to boosted regression trees. Journal of Animal Ecology 77(4), 802–813, doi: 10.1111/j.1365-2656.2008.01390.x Friedman, J. (1991), Multivariate adaptive regression splines. Annals of Statistics 19(1), 1–141. Hastie, T., Tibshirani, R., and A. Buja (1994), Flexible discriminant analysis by optimal scoring. Journal of the American Statistical Association 89(428), 1255–1270, doi: 10.1080/01621459.1994.10476866 Hastie, T., Buja, A., and R. Tibshirani (1995), Penalized discriminant analysis. The Annals of Statistics 23(1), 73–102. Hastie, T., and R. Tibshirani (1990), Generalized Additive Models, Monographs on Statistics and Applied Probability 43. Chapman and Hall, New York. McCullagh, P., and J. A. Nelder (1989), Generalized Linear Models. 2nd ed. Chapman & Hall, New York, 511pp. Phillips, S. J., Dudìk, M., and R. E. Schapire (2004), A maximum entropy approach to species distribution modeling. In: Proceedings of the 21st International Conference on Machine Learning, ACMPress, New York, pp. 655–662. Phillips, S. J., Anderson, R. P., and R. E. Schapire (2006), Maximum entropy modelling of species geographic distributions. Ecological Modelling 190(3–4), 231–259, doi: http://dx.doi.org/10.1016/j.ecolmodel.2005.03.026 Venables, W. N., and B. D. Ripley (2002), Modern Applied Statistics with S, fourth ed. Springer, Berlin, 495pp. Wood, S. N. (2011), Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. Journal of the Royal Statistical Society: Series B (Statistical methodology) 73(1), 3–36, doi: 10.1111/j.1467-9868.2010.00749.x