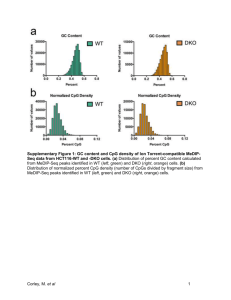
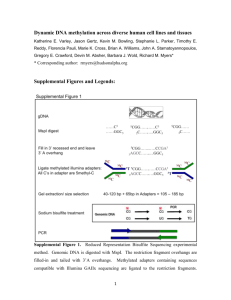
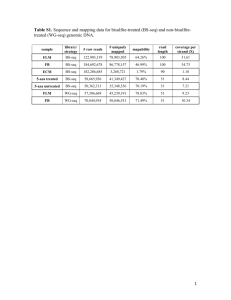
Supplementary Figure 1: overlap between fetal and colon cancer
advertisement

Supplementary Figure 1: overlap between fetal and colon cancer hypomethylated blocks. Black: proportion of fetal blocks at a given “area statistic” FWER cutoff that overlap the list of colon cancer blocks, red: proportion of overlapping blocks that are directionally consistent (lower DNAm levels in fetal and lower levels in cancer) among those blocks that overlap, blue line: cutoff corresponding to FWER ≤ 0.05 and overlap proportions reported in main text. Supplementary Figure 2: principal component analysis (PCA) of DNA methylation data, principal component 1 versus 2. Surrogate variables were estimated and regressed out of the methylation estimates prior to PCA and subsequent visualization. Samples are colored by age. Supplementary Figure 3: estimated neuronal composition versus age. Neuronal composition was estimated for each bulk tissue sample using flow sorted dorsolateral prefrontal cortex data (DLPFC) available from Guintivano et al (2013) as reference data and applying a statistical algorithm for cell composition deconvolution (Houseman et al, 2012; Montaño et al, 2013). The left-most panel shows the fetal samples, with x-axis values representing gestational age in weeks – other panels show age in years. The red line shows the linear spline (described in the Methods section) fit to the data. Supplemental Table 1: location of fetal DNA methylation “blocks”. Genomic coordinates are relative to genomic build hg19; “value” indicates the average difference in DNAm levels across the block – negative values mean fetal DNAm levels were lower than post-natal levels; “area” is the sum of the values within the block and the ranking metric; “L” and “clusterL” are the number of Illumina 450k probe groups and probes within the block, respectively; four measures of significance are provided – p.value and fwer (family-wise error rate) are based on the block “value” and p.valueArea and fwerArea are based on the block “area”, and were generated across 1000 permutations; “nearestCancerBlockDist” and “nearestCancerBlockSign” are the distance in base pairs to the nearest published colon cancer block, and its directionality (-1 means lower DNAm levels in cancer) from Hansen et al (2011) Supplemental Table 2: location and summary statistics of developmental DMRs. Genomic coordinates are relative to genomic build hg19; “value” indicates the average F-statistic for the region (see Methods section) ; “area” is the sum of the F-statistics values within the region and the ranking metric; “L” and “clusterL” are the number of Illumina 450k probes in the region and the number of probes in the probe group, respectively; three measures of significance are provided – fwer (family-wise error rate), pooled p-value and corresponding q-value (see Methods section and Jaffe et al (2012)) are based “area” statistics, and were generated across 500 permutations; the next 12 columns depict gene annotation information and can be generated using bumphunter::matchGenes() on this table. The next 6 columns depict the Pearson correlation and corresponding p-value for all, fetal, and post-natal samples, respectively, for the more correlated gene expression probe matching the gene symbol in the “name” column. Lastly, two additional gene identifiers are provided (Entrez Gene ID and RefSeq ID) based on the gene symbol. Regions with fwer ≤ 0.05 are considered genome-wide significant and discussed in the main text. References Guintivano J, Aryee MJ, Kaminsky ZA (2013) A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics : official journal of the DNA Methylation Society 8: 290-302 Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, Briem E, Zhang K, Irizarry RA, Feinberg AP (2011) Increased methylation variation in epigenetic domains across cancer types. Nature genetics 43: 768-775 Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT (2012) DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics 13: 86 Jaffe AE, Murakami P, Lee H, Leek JT, Fallin MD, Feinberg AP, Irizarry RA (2012) Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. International journal of epidemiology 41: 200-209 Montaño CM, Irizarry RA, Kaufmann WE, Talbot K, Gur RE, Feinberg AP, Taub MA (2013) Measuring cell-type specific differential methylation in human brain tissue. Genome biology 14: R94