Supplementary Information Figure 1 Figure 1: Wound healing assay
advertisement
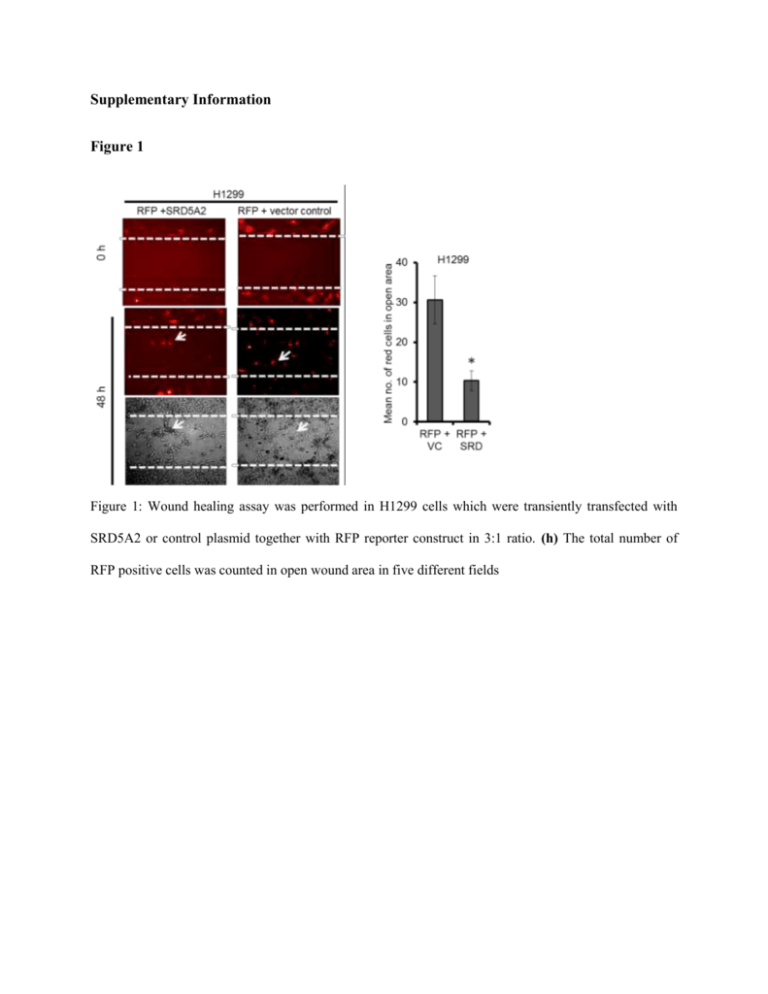
Supplementary Information Figure 1 Figure 1: Wound healing assay was performed in H1299 cells which were transiently transfected with SRD5A2 or control plasmid together with RFP reporter construct in 3:1 ratio. (h) The total number of RFP positive cells was counted in open wound area in five different fields Figure 2 Figure 2: LnCAP cells were transiently transfected with sh-SRD5A2 (SRD5A2 specific shDNA plasmid) or sh-Con (Control plasmid) alongwith pCMV-Gal expression vector (in 3:1 ratio). βGalactosidase activity was measured for transfection control using β-Gal assay kit (Promega). Figure 3. Figure 3: Cell growth property of S2, S4, VC and DU15 cells was examined by MTT assay. Cells were seeded in 96 well plates at 5000 cells/well. The 3-(4, 5- dimethylthiazol2yl) 2, 5 diphenyltetrazolium bromide (MTT) reagent was used at a final concentration of 1 ug/ul for the cell viability assay after 24, 48 and 72 hours. MTT reagent was added to the media and cells were incubated for 1 hour. The yellow MTT dye was reduced to insoluble purple formazan substrate which was solubilized in DMSO. The absorbance was measured at 570 nm. The experiment was performed in triplicate. Figure 4. Figure 4: Cell adhesion was examined in S2, S4, VC and DU145 cells. Cells grown after indicated time intervals were fixed and stained with crystal violet. Photomicrographs were taken under 10 X objective in bright field microscope. Figure 5. Figure 5: F-actin distribution was examined in S2, S4, VC and DU145 cells using PhalloidinFITC. Arrows show the actin stress fiber organisation in these cells. Figure 6. Figure 6. (a) S2 cells were targeted with sh-SRD or sh-Con plasmids along with reporter RFP vector (4:1). RFP positive cells were examined for actin distribution using phalloidin-FITC staining. (b) H1299 cells were transiently transfected with pBICEP-SRD5A2 or pBICEP vector along with H2Bmcherry reporter vector (4:1) and H2Bmcherry expressing cells were examined for actin distribution using phalloidin-FITC staining using two different filters. Figure 7. Fascin/ β-actin 2 1.6 1.2 0.8 0.4 0 S2 S4 VC DU145 Figure 7. Quantization of fascin protein bands (Fig 5 a, manuscript) is shown after normalizing with β-actin using Image J software. Figure 8. Figure 8: S2, S4, VC and DU145 cells were immunostained with mouse monoclonal anti-fascin antibody. TRITC labeled secondary antibody was used and cells were examined under Nikon E200 flourescent microscope. Figure 9. Figure 9. mRNA transcript levels of KRAS and Raf 1 genes were assessed in S2, S4, VC and DU145 cells by RT-PCR. GAPDH gene was used as internal control. Table 1 The gene specific primer sequences are listed in this table. Primer name Primer Sequence SRD5A2_F 5’ CATACGGTTTAGCTTGGGTG 3’ SRD5A2_R 5’ GCTTTCCGAGATTTGGGGTA 3’ MMP2_F 5’ TCTCCTGACATTGACCTTGGC 3’ MMP2_R 5’ CAAGGTGCTGGCTGAGTAGATC 3’ MMP7_F 5’ TGAGCTACAGTGGGAACAGG 3’ MMP7_R 5’ TCATCGAAGTGAGCATCTCC 3’ Raf1_F 5’ AATGAGCTTGCATGACTGCC 3’ Raf1_R 5’ AGGCAAGCTTCAGGAACGTG 3’ KRAS_F 5’ GTCTGCATGGAGCAGG 3’ KRAS_R 5’ AGAGGCCTGCTGAAAA 3’ β-actin_F 5’ TGCTATCCAGGCTGTGCTAT 3’ β-actin_R 5’ GATGGAGTTGAAGGTAGTTT 3’ Table 2. SRD5A2 Short haipin DNA (shDNA) containing pLKO.1 vectors were purchased from Sigma Aldrich (SHDNA 05080708MN). These were 5 vectors targeting different sites in the CDS of SRD5A2 mRNA. The details of sequence of each shRNA are provided in this table. The cocktail of all 5 shRNA plasmid DNA containing equal amount of each shDNA plasmid was used to silence SRD5A2 expression. TRCN0000026582 NM_000348.2-761s1c1 TRC 1 Region: CDS Alternate Species: NM_000348.2 Sequence: CCGGCCACCATAGGTTCTACCTCAACTCGAGTTGAGGTAGAACCTATGGTG GTTTTT TRCN0000026578 NM_000348.2-334s1c1 TRC 1 Region: CDS Alternate Species: NM_000348.2 Sequence: CCGGGCCTACATTACTTCCACAGGACTCGAGTCCTGTGGAAGTAATGTAGG CTTTTT TRCN0000026553 NM_000348.2-458s1c1 TRC 1 Region: CDS Alternate Species: NM_000348.2 Sequence: CCGGTCTGATTTACTGTGCTGAATACTCGAGTATTCAGCACAGTAAATCAGA TTTTT TRCN0000026548 NM_000348.2-776s1c1 TRC 1 Region: CDS Alternate Species: NM_000348.2 Sequence: CCGGCCTCAAGATGTTTGAGGACTACTCGAGTAGTCCTCAAACATCTTGAG GTTTTT TRCN0000026506 NM_000348.2-631s1c1 TRC 1 1 Region: CDS Alternate Species: NM_000348.2 Sequence: CCGGCGTATGTTTCTGGAGCCAATTCTCGAGAATTGGCTCCAGAAACATAC GTTTTT