Key Concepts Lectures 13-15
advertisement

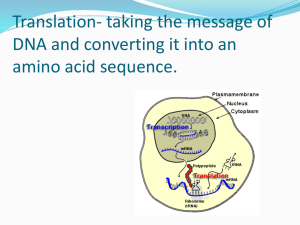
Key Concepts for Week 5: Lectures 13-15 Monday, Oct. 26, Lecture 13: (Transcription and) Translation DNA mRNA (occurs in the nucleus) Initial DNA strand used to create primary transcript (mRNA) Codon: three nucleotide sequence that codes for an amino acid (20 AAs) – triplet of nucleotides is the smallest unit to code for all AAs o 64 codons total with 3 being STOP codons to end translation Template strand: one DNA strand that is designated for transcription Steps of transcription: o Initiation Promoter region important for initiating transcription Transcription factors bring in machinery needed for starting transcription (e.g. allowing binding of RNA polymerase) Transcription initiation complex o Elongation DNA double helix untwists as RNA polymerase moves and transcribes strand o Termination 5’ end gets a nucleotide cap (G-cap) 3’ end gets a poly A tail Modifications at ends of strand help protect strand from degradation and allows transport of mRNA out into nucleus for translation mRNA protein (occurs in the cytoplasm) tRNA – transfer RNA used to translate mRNA message into a protein o Each tRNA has an anticodon that pairs with the complementary codon on mRNA Aminoacyl-tRNA synthetase matches a specific amino acid with its particular tRNA Ribosomes specifically couple tRNA anticodons with mRNA codons – ribosomes have 3 sites o P site: holds tRNA that has the polypeptide chain o A site: holds tRNA that holds the next amino acid that needs to be added o E site: exit site where tRNAs leave Steps of translation o Initiation: start codon (AUG) begins translation o Elongation Amino acids are running one by one to the preceding amino acid tRNA shifts from P – E ribosomal site until it hits a stop codon o Termination When stop codon reaches A site of ribosome, A site accepts a protein called a release factor Release factor adds H20 onto peptide instead of amino acid Polyribosomes o Multiple ribosomes that can perform translation simultaneously, forming a polyribosome (efficient translation machinery) Creating a functional protein requires modification following translation o Specific proteins need to go to specific areas in the cell o Proteins in the cytosol mostly made by free ribosomes o Proteins secreted or used in endomembrane system made by bound ribosomes (review of earlier lecture series) o Signal recognition particle (SRP): signal telling where a peptide belongs Gene mutations o Point mutations Nucleotide pair substitutions Silent mutations Missense mutations Nonsense mutations Nucleotide pair insertions/deletions Wednesday, Oct. 28, Lecture 14: ATP, Enzymes What is ATP? o Used for mechanical and chemical work o Structure of ATP Endergonic and exergonic reactions o Exergonic: Reaction that releases energy, occurs spontaneously Hydrolysis of ATP ADP + P (releases energy and requires an enzyme, ATPase) o Endergonic: Reaction that takes in energy, occurs nonspontaneously Energy used towards converting ADP + P ATP Enzymes o Catalysts – speed up chemical reactions by lowering energy barrier o Have high substrate specificity – each enzyme has an active site that substrate binds onto, causing a change in enzyme shape Friday, Oct. 30, Lecture 15: Cellular Respiration Redox reactions o Mechanism behind cellular respiration Oxidation: loss of electrons Reduction increase of electrons “OIL RIG”: oxidation is losing electrons, reduction is gaining electrons Mitochondria is where cellular respiration takes place o O2 + C6H12O6 (glucose) CO2 + H2O and energy O2 gains a H+ (and electron), to make H2O is reduced Glucose loses H+ (and electron), to make CO2 is oxidized Cellular respiration (3 steps) – outlined in Fig 9.6 o Glycolysis Glucose pyruvate (occurs in cytosol) Basic way to generate ATP (2 ATP) o Pyruvate oxidation and the citric acid cycle Pyruvate goes into mitochondrial matrix and gets converted into acetyl CoA Acetyl CoA conversion generates NADH and FADH2 that goes to electron transport chain 2 ATP for every turn of cycle o Oxidative phosphorylation: electron transport and chemiosmosis Majority of ATP generated here (32 ATP) Electron transport chain NADH and FADH2 donate electrons energy exchange yields H20 Controlled release of energy Pumps H+ across membrane Chemiosmosis H+ gradient provides basis for cell to make ATP