tpj12292-sup-0021-TableS5
advertisement
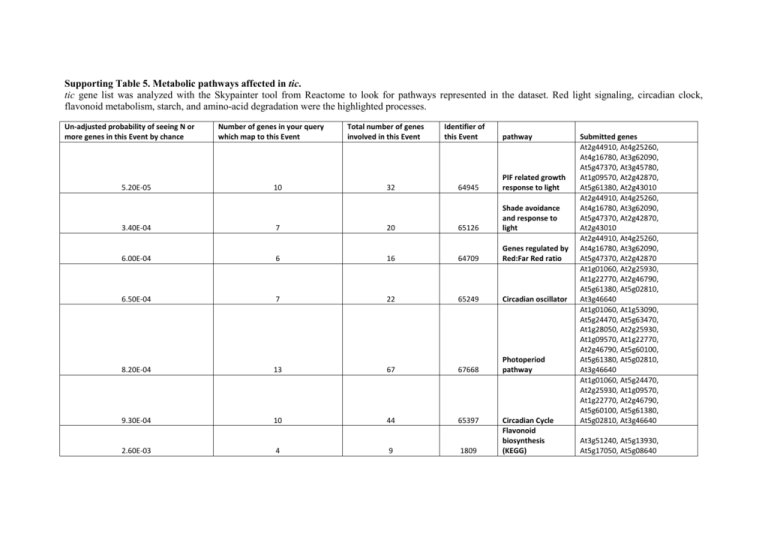
Supporting Table 5. Metabolic pathways affected in tic. tic gene list was analyzed with the Skypainter tool from Reactome to look for pathways represented in the dataset. Red light signaling, circadian clock, flavonoid metabolism, starch, and amino-acid degradation were the highlighted processes. Un-adjusted probability of seeing N or more genes in this Event by chance 5.20E-05 Number of genes in your query which map to this Event 10 Total number of genes involved in this Event 32 Identifier of this Event pathway 64945 PIF related growth response to light 3.40E-04 7 20 65126 Shade avoidance and response to light 6.00E-04 6 16 64709 Genes regulated by Red:Far Red ratio 6.50E-04 7 22 65249 Circadian oscillator 8.20E-04 13 67 67668 Photoperiod pathway 9.30E-04 10 44 65397 2.60E-03 4 9 1809 Circadian Cycle Flavonoid biosynthesis (KEGG) Submitted genes At2g44910, At4g25260, At4g16780, At3g62090, At5g47370, At3g45780, At1g09570, At2g42870, At5g61380, At2g43010 At2g44910, At4g25260, At4g16780, At3g62090, At5g47370, At2g42870, At2g43010 At2g44910, At4g25260, At4g16780, At3g62090, At5g47370, At2g42870 At1g01060, At2g25930, At1g22770, At2g46790, At5g61380, At5g02810, At3g46640 At1g01060, At1g53090, At5g24470, At5g63470, At1g28050, At2g25930, At1g09570, At1g22770, At2g46790, At5g60100, At5g61380, At5g02810, At3g46640 At1g01060, At5g24470, At2g25930, At1g09570, At1g22770, At2g46790, At5g60100, At5g61380, At5g02810, At3g46640 At3g51240, At5g13930, At5g17050, At5g08640 5.30E-03 2 2 42410 5.30E-03 5.30E-03 2 2 2 2 42959 63732 5.30E-03 2 2 63953 5.30E-03 2 2 64487 5.30E-03 2 2 66556 5.30E-03 2 2 67953 6.50E-03 3 6 67266 7.70E-03 5 18 32318 8.60E-03 4 12 20299 1.20E-02 4 13 68868 1.50E-02 1.50E-02 2 2 3 3 14477 17865 methionine degradation II (AraCyc) L-methionine + H2O <=> NH3 + 2oxobutanoate + methanethiol PRR3 Binds TOC1 COL15 interacts with the HAP5 protein AT5G63470 PIF6 interacts with TOC1 PIF4 interacts with GAI PIF4 interacts with TOC1 Interaction between photomorphogensi s and the circadian clock starch degradation (AraCyc) arginine degradation I (AraCyc) Evening Loop 3 H2O + GTP <=> pyrophosphate + 2,5-diamino-6(ribosylamino)-4(3H)-pyrimidinone 5'-phosphate + formate D-ribulose-5- At1g77670, At1g64660 At1g77670, At1g64660 At5g60100, At5g61380 At5g63470, At1g28050 At3g62090, At5g61380 At2g44910, At2g43010 At5g61380, At2g43010 At3g62090, At5g61380, At2g43010 At4g17090, At5g18670, At5g55700, At3g52180, At1g69830 At2g16500, At3g08860, At1g79440, At2g38400 At1g01060, At2g25930, At1g22770, At3g46640 At5g59750, At2g22450 At5g59750, At2g22450 1.50E-02 4 14 18808 phosphate <=> 3,4dihydroxy-2butanone-4-P + formate biotin biosynthesis II (AraCyc) chlorophyllide a biosynthesis (AraCyc) 2.30E-02 9 58 32887 2.30E-02 3 9 63432 2.90E-02 2 4 25592 2.90E-02 2 4 69427 3.10E-02 4 17 66281 3.10E-02 3 10 20193 3.10E-02 3 10 27507 3.10E-02 3 10 65764 4.60E-02 2 5 14252 Morning Loop D-fructose-6phosphate + UDPD-glucose <=> sucrose-6'phosphate + UDP UDP-D-Glucose + DFructose 6phosphate => Sucrose 6phosphate + UDP Starch degradation in leaves Long-linear-glucans <=> maltose glutamate degradation II (AraCyc) Seedling deetiolation L-alanine + 6carboxyhexanoylCoA <=> CO2 + coenzyme A + 8amino-7oxononanoate At4g36480, At3g08860, At1g08630, At2g38400 At5g18660, At3g14930, At2g43020, At4g03140, At5g54190, At3g08860, At2g38400, At5g02540, At4g27440 At1g01060, At2g46790, At5g02810 At5g17310, At5g20280 At5g17310, At5g20280 At4g17090, At1g10760, At3g29320, At1g69830 At4g17090, At5g18670, At5g55700 At3g08860, At1g79440, At2g38400 At3g45780, At1g09570, At2g43010 At4g36480, At1g08630 4.60E-02 2 5 16699 4.60E-02 2 5 25067 4.60E-02 2 5 32167 4.60E-02 2 5 34032 Long-linear-glucans <=> Short-glucans glutamate-1semialdehyde <=> 5-amino-levulinate glutamate-1semialdehyde <=> 5-amino-levulinate Starch <=> Largebranched-glucans At3g52180, At1g69830 At3g08860, At2g38400 At3g08860, At2g38400 At3g52180, At1g69830