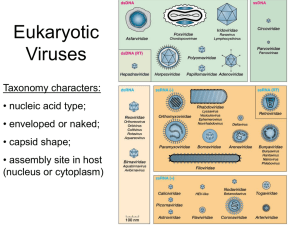
Chapter 3: Genomes Types of genomes RNA or DNA RNA viruses
advertisement

Chapter 3: Genomes Types of genomes RNA or DNA o RNA viruses must bring/encode a RNA-dependent RNA polymerase o DNA viruses can use host replication machinery (DNA-dependent RNA polymerase and DNA polymerase) or encode their own Single stranded (ss) or double stranded (ds) Positive sense (+), negative sense (-), or ambisense (mixture of – and +) o + sense genomes can be directly translated into protein +ssRNA viruses can translate an RNA-dependent RNA polymerase after entering the cell o – sense must first be transcribed into + mRNA then made into protein -ssRNA viruses must carry a virion-associated RNA-dependent RNA polymerase with them (packaged in the virion) in order to produce a + strand upon entering the cell Can be segmented Monocistronic mRNA problem: most viruses have just one strand of genomic material which encodes multiple proteins. But the host cell is used to having one mRNA for one protein. Viral mechanisms to get around this problem: Internal ribosome entry sites Make polyproteins and then cleave them into multiple proteins Alternative splicing to make multiple mRNAs from a single strand Mutations Strain: different lines/isolates of a virus Type: different serotypes (an antibody neutralization phenotype—viruses of the same serotype are neutralized by the same antibodies) Variant: a virus that differs from wild-type strain but the genetic difference is unknown Mutations arise due to errors in the genome replication o Most viral replication machinery doesn’t have error checking mechanisms like host replication machinery Mutations can be induced by chemicals Mutated viruses can go back to a wild-type phenotype via a second mutation o Back mutation (undoing) of the original mutation o Compensatory mutation in the same gene as the original mutation (intragenic suppression) o Suppressor mutation in a different gene (extragenic mutation) Two mutated viruses can complement each other if they have mutations in different complementation groups o Requires co-infection by both viruses o Ex: one virus has a mutation that results in an un-functional protein. The second virus produces functional protein and thus the first virus can use it and be infective. Transposition o Transposable genetic elements move from one position in the genome to another Simple ones are found in prokaryotes (e.g. phages) and do not undergo reverse transcription (RNA to DNA) Retrotransposons are bounded by long direct repeats and undergo reverse transcription to jump around the genome Retroviruses are essentially a retrotransposon with an envelope protein Recombination Two viruses infecting the same cell can undergo recombination, resulting in different genomes and possibly altered infectivity Reassortment Only in viruses with segmented genomes The segments get shuffled up during virus production If two viruses infect the same cell, then the segments can get shuffled between them (e.g. flu viruses) Phenotypic Mixing Two viruses infecting the same cell Viral genomes may be packed with proteins (e.g. capsids) from the other virus A few viruses… Large dsDNA genomes: adenoviruses and herpesviruses o Adenoviruses use alternative splicing to make multiple mRNA and a terminal protein during replication Parvoviruses: ssDNA, contains only two genes, the end of the genomes fold over to form hairpins which initiate replication Polyomaviruses: dsDNA, supercoiled and associated with histones. + ssRNA viruses o Fragile, high mutation rates, hence they have smaller genomes than DNA viruses o Picornaviruses: +ssRNA, non enveloped. Use an internal ribosome entry site and make a polyprotein which is then cleaved into smaller proteins. o Togaviruses: use a 5’ methylated cap and 3’ poly(A) sequence on its RNA to mimic cellular mRNA. Produce non-structural proteins before structural proteins o Flaviviruses: use a 5’ methylated cap but are not polyadenylated, produce a polyprotein o Coronaviruses: use a 5’ cap and polyA tail. First makes a RNA polymerase, which then produces mRNA as a “nested” set of transcripts (discontinuous subgenomic transcription). -ssRNA viruses: more diverse than +RNA viruses, have larger genomes. Segmentation is common. Do not use 5’ capping or 3’ polyadenylation. o Bunyaviruses: segmented, some are ambisense (5’ end is + but 3’ end is – sense) o Also arenaviruses (segmented), paramyxoviruses, and rhabdoviruses Segmented viruses o Have less strand breakage since they have shorter genomes o Must package all segments into virus particle o Multipartite genomes are segmented but each segment is in a separate particle o Can undergo reassortment—rapid generation of genetic diversity o Influenza is the model reassortment virus! Retroviruses o The only viruses that are diploid o RNA genome is turned into DNA with viral reverse transcriptase and then integrated into the host genome by viral integrase protein. The DNA genome is longer than the RNA due to duplication of long terminal repeats at the end of the genome Reverse transcriptase is highly error-prone!! Rapid genetic variation arises! o Only RNA viruses whose genome is made by host transcriptional machinery (DNA-dependent RNA polymerase) Virus Evolution o Regressive: viruses could be degenerate life-forms which have lost many functions of other organisms and are now parasites o Cellular origins: viruses are subcellular assemblies of macromolecules that have escaped from their cellular origins o Independent entities: evolved on a parallel course to cellular organisms from primitive self-replicating molecules in the “RNA world” Chapter 4: Replication Ellis and Delbruck experiment o Showed three essential phases of virus replication via bacteriophages infecting rapidly growing bacteria Initiation of infection Replication and expression of virus genome Release of mature virions from cell Hershey-Chase experiment o Used radioisotopes that were incorporated into proteins and nucleic acids o Showed that nucleic acid (and not protein) carried the genetic information of phages and viruses General replication cycle o Attachment by interaction between virus protein and cellular receptor molecule Virus receptors include immunoglobulin-like molecules, membraneassociated receptors, transmembrane transporters channels Ex. Flu viruses use hemagglutinin to attach to cell receptors and neuraminidase to cleave that attachment—this releases the virus from inappropriate binding (e.g. to cellular debris). Virus and cell receptors determine the tropism of a virus. Tropism means the type of host cell that the virus can replicate in. This influences viral pathogenesis (the disease that the virus causes to the host/cell) Some viruses require 2 receptor interactions to enter a cell (e.g. HIV and adenoviruses) o Penetration Fusion of envelope with plasma membrane Translocation of entire virus particle across plasma membrane— relatively rare mechanism among viruses, mediated by proteins in virus capsid and cell membrane receptors Receptor mediated endocytosis o Uncoating of genome-poorly understood, depends on structure of nucelocapsid o Expression of proteins dsRNA—all these viruses have segmented genomes each segment transcribed separately to produce monocistronic mRNAs dsDNA use host cell machinery to make mRNA (except poxviruses which use viral machinery) +ssRNA Some have polycistronic mRNA, which is then made into a polyprotein and cleaved into individual proteins Some use complex transcription to produce monocistronic mRNA o Genome replication dsDNA o o o o replication occurs in the nucleus for all viruses except poxviruses, which have the necessary genes for transcription and replication of their genomes so they don’t need host cell machinery ssDNA form a double-stranded intermediate, replicate in the nucleus RNA viruses Replicate in cytoplasm (except orthomyxoviruses, which includes the flu) Retroviruses + diploid RNA, replicate via a DNA intermediate that is integrated into the host genome dsDNA viruses with an RNA intermediate rely on reverse transcriptase, but this reverse transcription occurs inside the virus particle during maturation Assembly All components necessary for the formation of the mature virion must collect at a particular site in the cell Site of assembly depends on site of replication in the cell and release mechanism Maturation of virions A mature virus is an infectious virus Involves structural changes in the virus particle (e.g. cleavage of capsid proteins by viral proteases resulting in conformational changes) Release of virions Cell lysis Most non-enveloped viruses are released by this route The infected cell breaks open and releases the viruses Budding Enveloped viruses acquire their envelope (lipid membrane) as the virus buds out of the cell through the cell membrane or Golgi apparatus Bud out of the cell through the cell membrane or are released from an intracellular vesicle