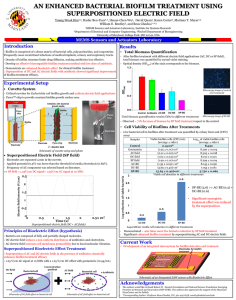
bit25867-sup-0001-SuppData
advertisement

1
Supplementary materials
2
The method descrided here are described in detail by Li et al. (2015), for more
3
detailed information, please refer to their publication.
4
Estimation of evaporation rate and effective diffusion coefficient in biofilm
5
The porosity of the biofilm (θ) was estimated from the volume of cells (biovolume) in
6
the biofilm divided by the total biofilm volume. The biovolume was calculated as
7
described by Hillebrand et al. (1999) from suspended biofilm samples with a 50 μm
8
resolution by means of a custom-made hand microtome (Fig. S2). The biofilm
9
immobilized on a polycarbonate membrane was placed onto a height-adjustable stage
10
on a thin layer of culture medium. The stage allows levelling the biofilm sample into
11
the sectioning plane by means of a micrometer screw. The sectioning plane was
12
defined by a solid brass support over which a rigid microtome blade was passed
13
manually to remove the overlying algal biofilm (Fig. S2).
14
Evaporation rate during the measurement was 0.68 mL min-1 (monitored by recording
15
the change of the volume in the medium container, Fig S1), which corresponded to a
16
convective flow rate of 5.4 µm s-1 inside the biofilm, which was calculated as:
18
𝑢 = 𝑞𝑒 /(𝐴𝑒 ∙ 𝜃)
17
19
(Eq.S1),
qe is the rate of liquid volume lost, Ae is the exposed surface area of the biofilm.
1
20
Microsensor setup
21
A schematic representation of the experimental setup for microsensor measurements
22
based on previous works by Gieseke and de Beer (2004) is given in Fig. S1: the
23
biofilm immobilized on a polycarbonate membrane was placed onto a moist glass
24
fiber inside the biofilm measurement cell under a controlled atmosphere of
25
compressed air at a flow rate of 1 L min-1 (Fig. S1A). The non-inoculated area of the
26
polycarbonate filter and glass fiber were covered with a black plastic foil to exclude
27
light and gas exchange in this area. 50 mL of BBM culture medium were circulated
28
though the measurement cell with a flow rate of 3 mL min-1 by means of a peristaltic
29
pump and were exchanged every 1 hour during the measurement. Light was supplied
30
from the front side by a halogen lamp (KL-1500, Schott, Mainz, Germany) equipped
31
with a 3-fold splitter to ensure even distribution. The movement of the sensor was
32
enabled by a computer-controlled micromanipulator (Pollux Drive, PI miCos,
33
Eschbach, Germany). Microsensor signals were amplified and converted into digital
34
data (DAQpad 6015 and 6009, National Instruments, Munich, Germany) prior to their
35
further processing on a computer (Fig. S1B). Software used for system control and
36
data acquisition was custom made (can be acquired upon request from Dr. Lubos
37
Polerecky, Utrecht University, Netherlands, l.polerecky@uu.nl). Light-dark shift
38
measurements (Revsbech, 1989) were carried out, and the data acquired was
39
processed as described in the following section. Linear calibration of oxygen
40
microsensor was carried out as described by Revsbech (1983) using BBM medium
2
41
saturated with nitrogen gas or compressed air. A shutter connected to a timer was
42
used to control light-dark cycling: the total dark period was 3 s, and a linear
43
regression slope of oxygen concentration change between 0.7 and 2.1 s of the dark
44
period was determined as the measured value. For each measurement depth, three
45
light-dark cycles were performed after steady state was reached (this could be as long
46
as 10 min), and these 3 values acquired were considered as triplicates. The PAR light
47
intensity of a fixed depth was determined as the average value of a 3 s measurement
48
period.
49
Gross photosynthetic productivity calculation
50
Take the derivative of time on both sides of the dissolved oxygen mass transfer
51
equation, and assume the time t and spatial variables x are not dependent (as during
52
the measurement period, the thickness of the biofilm hardly changes) (Bergman et al.,
53
2011, Revsbech and Jorgensen, 1983):
55
𝜕𝐶
𝜕𝐶
𝜕2 ( )
𝜕( )
𝜕𝐶
𝜕𝑡 + 𝑢
𝜕𝑡 − 𝜕𝑅 /𝜕𝑡
𝜕( )/𝜕𝑡 = 0/𝜕𝑡 − 𝜕𝑅𝑙 /𝜕𝑡 + 𝐷𝑒
𝑠
𝜕𝑡
𝜕𝑥 2
𝜕𝑥
54
(Eq.S2).
56
The term ∂Rs/∂t represents the rate change of the removal of dissolved gaseous
57
species at the submerged biofilm surface due to the flow of bulk liquid, or, in this
58
case, of the gas phase above the biofilm:
59
𝜕𝑅𝑠 /𝜕𝑡 = (𝐷𝑒
(𝐶 − 𝐶𝑠 )
(𝐶 − 𝐶𝑠 )
+
𝑢
)/𝜕𝑡
𝑥2
𝑥
3
=(
61
𝐷𝑒 𝑢
+ ) ∙ 𝜕𝐶/𝜕𝑡
𝑥2 𝑥
60
(Eq.S3).
62
Cs represents the oxygen concentration on the very surface of the aerial biofilm,
63
which is in equilibrium with the phase above and is considered to be a constant.
64
Considering that the rate of respiration remains stable during the measurement period
65
(Glud et al., 1992), and take ∂C/∂t=P(x, t), as ∂C/∂t is a function of depth x and time t.
66
Substitute Eq.S3 into ∂Rs/∂t and ∂C/∂t to P(x, t), Eq.S2 becomes:
68
𝜕 2 (𝑃(𝑥, 𝑡))
𝜕(𝑃(𝑥, 𝑡))
𝜕𝑃(𝑥, 𝑡)
𝐷𝑒 𝑢
= 0 − 0 + 𝐷𝑒
+𝑢
− ( 2 + ) ∙ 𝑃(𝑥, 𝑡)
2
𝜕𝑡
𝜕𝑥
𝜕𝑥
𝑥
𝑥
67
(Eq.S4).
69
Eq.S4 is a nonhomogeneous diffusion equation with a sink and can be solved by using
70
any of the methods summarized by Crank or Polyanin (Crank, 1975; Polyanin, 2002).
71
Using an initial condition P(x, t = 0) = P(x, 0) and a boundary condition ∂P(x = 0,
72
t)/∂t = 0, the exact solution of Eq.S4 at t = τ can be expressed as:
𝛿
74
𝑃(𝑥, 𝜏) = ∫ 𝑃(𝑦, 0) ∙ [𝑒
−(𝑦−𝑥+𝑢∙𝜏)
(
)
4∙𝐷𝑒 ∙𝜏
0
−𝑒
−(𝑦+𝑢∙𝜏)
(
)
4∙𝐷𝑒 ∙𝜏
∙ 𝑒𝑟𝑓𝑐(
𝑥
√4 ∙ 𝐷𝑒 ∙ 𝜏
)] ∙ 𝑑𝑦
73
(Eq.S5),
75
δ is the total depth of the biofilm, or, alternatively, the depth until which the shift from
76
light to dark has an effect on the dissolved oxygen concentration. P(x, τ) is the
77
measured value from the light-dark shift method with measurement time τ at
78
measurement depth x, and P(y, 0) is the actual photosynthetic activity at position y
4
79
(the integrating variable), which is the desired term. For measurements taken at all
80
depths, the measured value P(x, τ) at a depth x depends on the P(y, 0) across the
81
complete photosynthetically active region of the biofilm (surface to depth δ).
82
Eq.S5 is a Fredholm integral equation of the first kind (Kress, 2014), and its
83
approximated solution can be calculated using Tikhonov regularization (Tikhonov et
84
al., 1995) with a non-negative constrain applying the active-set algorithm purposed by
85
Gill et al. (1981): Discrete δ by n, and let P(xj, τ) = gj, P(yi, 0) = hi; (i, j = 1, 2, …, n)
86
and write Eq.S5 in the discrete operator notion.
𝑛
𝑔𝑗 = ∑ 𝑘𝑖,𝑗 ∙ ℎ𝑖
88
𝑖
87
(Eq.S6),
89
ki,j is the operator term which contains both the term that reflects the effect of
90
diffusion and advection of dissolved oxygen inside the biofilm (inner term, first term
91
in square bracket in Eq.S5) and the term that reflects the effect of dissolved oxygen
92
removal due to surface flow (surface term, second term in square bracket in Eq.S5). In
93
matrix notion, let G be a vector containing gj, H a vector containing hi, and K a matrix
94
containing the operator terms ki,j:
𝐺 =𝐾∙𝐻
96
95
97
(Eq. S7).
Rewrite Eq.S7 as:
5
(𝐺𝑖𝑛 + 𝐺𝑠𝑢𝑟𝑓 ) = (𝐾𝑖𝑛 + 𝐾𝑠𝑢𝑟𝑓 ) ∙ 𝐻
99
98
(Eq.S9),
100
The subscripts in and surf represent the effect from the inner term and the surface
101
term in Eq.S5 respectively, operators Kin, Ksurf can be calculated using Eq.S5, as well
102
as gin and Gsurf, if an H is given. Apply the Tikhonov regularization, the problem in
103
Eq.11 thus becomes:
2
min {‖(𝐺𝑖𝑛 + 𝐺𝑠𝑢𝑟𝑓 ) − (𝐾𝑖𝑛 + 𝐾𝑠𝑢𝑟𝑓 ) ∙ 𝐻‖ + 𝜆2 ∙ ‖𝐿 ∙ 𝐻‖2 }
104
105
107
𝐹≥0
or,
2
min {‖(𝐺𝑖𝑛 − 𝐾𝑖𝑛 ∙ 𝐻) + (𝐺𝑠𝑢𝑟𝑓 − 𝐾𝑠𝑢𝑟𝑓 ∙ 𝐻)‖ + 𝜆2 ∙ ‖𝐿 ∙ 𝐻‖2 }
𝐹≥0
106
(Eq.S10).
108
Observing the surface term in Eq.S5, notice the maximum value of the surface term is
109
controlled by the position of the actual activity y, but a complement error function
110
dependent solely on the position of the measurement taken (x) controls the final value.
111
Furthermore, for a given G, under the same measurement conditions, Gsurf always has
112
the same set of values. Thus, for a given G, the minimization of the term
113
‖𝐺𝑠𝑢𝑟𝑓 − 𝐾𝑠𝑢𝑟𝑓 ∙ 𝐻‖ will always yield the same solution. As a result, the problem in
114
Eq. S10 can be simplified to:
116
2
min{‖𝐺𝑖𝑛 − 𝐾𝑖𝑛 ∙ 𝐻‖2 + 𝜆2 ∙ ‖𝐿 ∙ 𝐻‖2 }
𝑓≥0
115
(Eq. S11).
6
117
Eq.S11 does not contain the surface term, but the solution of the problem still leads to
118
H, which is the desired original profile. The L-curve method purposed by Hansen &
119
O'Leary (1993) was used to find the best regularization parameter λ.
120
The treatment procedure was coded in MATLAB (version 2013a, MathWorks,
121
Ismaning, Germany), and the inbuilt quadprog was used for solving the minimization
122
problems. L-curve function from the regularization tools (Hansen, 1994) was used to
123
find the λ value. The mean values of triplicates acquired from the microsensor
124
measurement were randomly added or subtracted with a random value in range of the
125
calculated standard deviation (SD) at the same position. These values were used as
126
input for the calculation. The calculation procedure was repeated 3 times, and the
127
mean value of the 3 results was taken as the final result (a SD can also be calculated).
128
Data interpolation, if needed, was done by simply connecting two measured data
129
points with a straight line (linear interpolation, using MATLAB inbuilt function
130
interp1).
7
131
References for supplementary materials:
132
Bergman TL, Lavine AS, Incropera FP, DeWitt DP. 2011. Fundamentals of heat and
133
mass transfer. 7th ed. Hoboken, New Jersey, USA: John Wiley & Sons. 1048 p.
134
Crank J, editor. 1975. Mathematics of diffusion. 2nd ed. Oxford, UK: Oxford
135
University Press. 414 p.
136
Gieseke A, de Beer D. 2004. Use of microelectrodes to measure in situ microbial
137
activities in biofilms, sediments, and microbial mats. In: Kowalchuk GA, de Bruijn
138
FJ, Head IM, Akkermans AD, van Elsas JD, editors. Molecular Microbial Ecology
139
Manual. The Netherlands: Springer. p 3483-3514.
140
Gill PE, Murray W, Wright MH. 1981. Practical optimization. Waltham,
141
Massachusetts, USA: Academic Press. 401 p.
142
Glud RN, Ramsing NB, Revsbech NP. 1992. Photosynthesis and photosynthesis-
143
coupled respiration in natural biofilms quantified with oxygen microsensors. Journal
144
of Phycology 28(1):51-60.
145
Hansen P. 1994. REGULARIZATION TOOLS: A Matlab package for analysis and
146
solution of discrete ill-posed problems. Numerical Algorithms 6(1):1-35.
8
147
Hansen PC, O’Leary DP. 1993. The Use of the L-Curve in the Regularization of
148
Discrete Ill-Posed Problems. SIAM Journal on Scientific Computing 14(6):1487-
149
1503.
150
Hillebrand H, Durselen CD, Kirschtel D, Pollingher U, Zohary T. 1999. Biovolume
151
calculation for pelagic and benthic microalgae. Journal of Phycology 35(2):403-424.
152
Kress R, editor. 2014. Linear integral equations. 3rd ed. New York City, USA:
153
Springer. 412 p.
154
Li T, Podola B, de Beer D, Melkonian M. 2015. A method to determine
155
photosynthetic activity from oxygen microsensor data in biofilms subjected to
156
evaporation. Journal of Microbiological Methods 117:100-107.
157
Polyanin AD. 2002. Handbook of linear partial differential equations for engineers
158
and scientists. Boca Raton, Florida: Chapman&Hall/CRC Press. 800 p.
159
Revsbech NP. 1983. In Situ Measurement of Oxygen Profiles of Sediments by use of
160
Oxygen Microelectrodes. In: Gnaiger E, Forstner H, editors. Polarographic Oxygen
161
Sensors. Berlin/Heidelberg, Germany: Springer. p 265-273.
9
162
Revsbech NP, Jorgensen BB. 1983. Photosynthesis of benthic microflora measured
163
with high spatial-resolution by the oxygen microprofile method - capabilities and
164
limitations of the method. Limnology and Oceanography 28(4):749-756.
165
Tikhonov AN, Goncharsky A, Stepanov VV, Yagola AG. 1995. Numerical methods
166
for the solution of ill-posed problems. Dordrecht, the Netherlands: Springer. 254 p.
Supplementary materials figure captions
167
Figure S1
168
Experimental setup for microsensor studies in non-submerged microalgal biofilms. A:
169
Schematic drawing of the vertical cross section of the measurement in artificial
170
biofilms. Solid arrows in the glass fiber tissue indicate flow direction of the culture
171
medium. B: Schematic drawing of the complete experimental setup, dotted lines with
172
arrow indicate signal flow, whereas solid arrows represent medium and gas flows in
173
the system.
174
Figure S2
175
Schematic drawing of the experimental setup used for biofilm sectioning. The sample
176
holder allows moving the biofilm in vertical direction by means of a micrometer
177
screw (indicated by the thin solid arrows). The biomass above the sectioning plane
10
178
was removed with a solid microtome blade. The cutting direction is indicated by the
179
thick arrow.
11