tpj12208-sup-0005-TableS2
advertisement
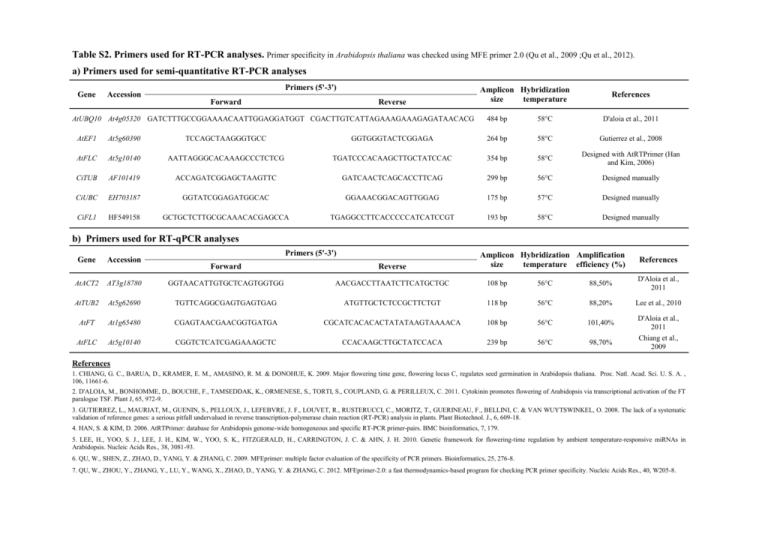
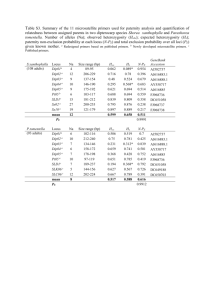
Table S2. Primers used for RT-PCR analyses. Primer specificity in Arabidopsis thaliana was checked using MFE primer 2.0 (Qu et al., 2009 ;Qu et al., 2012). a) Primers used for semi-quantitative RT-PCR analyses Gene Primers (5'-3') Accession Forward Reverse AtUBQ10 At4g05320 GATCTTTGCCGGAAAACAATTGGAGGATGGT CGACTTGTCATTAGAAAGAAAGAGATAACACG Amplicon Hybridization size temperature References 484 bp 58°C D'aloia et al., 2011 AtEF1 At5g60390 TCCAGCTAAGGGTGCC GGTGGGTACTCGGAGA 264 bp 58°C Gutierrez et al., 2008 AtFLC At5g10140 AATTAGGGCACAAAGCCCTCTCG TGATCCCACAAGCTTGCTATCCAC 354 bp 58°C Designed with AtRTPrimer (Han and Kim, 2006) CiTUB AF101419 ACCAGATCGGAGCTAAGTTC GATCAACTCAGCACCTTCAG 299 bp 56°C Designed manually CiUBC EH703187 GGTATCGGAGATGGCAC GGAAACGGACAGTTGGAG 175 bp 57°C Designed manually CiFL1 HF549158 GCTGCTCTTGCGCAAACACGAGCCA TGAGGCCTTCACCCCCATCATCCGT 193 bp 58°C Designed manually b) Primers used for RT-qPCR analyses Gene Accession AtACT2 Primers (5'-3') Amplicon Hybridization Amplification size temperature efficiency (%) References Forward Reverse AT3g18780 GGTAACATTGTGCTCAGTGGTGG AACGACCTTAATCTTCATGCTGC 108 bp 56°C 88,50% D'Aloia et al., 2011 AtTUB2 At5g62690 TGTTCAGGCGAGTGAGTGAG ATGTTGCTCTCCGCTTCTGT 118 bp 56°C 88,20% Lee et al., 2010 AtFT At1g65480 CGAGTAACGAACGGTGATGA CGCATCACACACTATATAAGTAAAACA 108 bp 56°C 101,40% D'Aloia et al., 2011 AtFLC At5g10140 CGGTCTCATCGAGAAAGCTC CCACAAGCTTGCTATCCACA 239 bp 56°C 98,70% Chiang et al., 2009 References 1. CHIANG, G. C., BARUA, D., KRAMER, E. M., AMASINO, R. M. & DONOHUE, K. 2009. Major flowering time gene, flowering locus C, regulates seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. , 106, 11661-6. 2. D'ALOIA, M., BONHOMME, D., BOUCHE, F., TAMSEDDAK, K., ORMENESE, S., TORTI, S., COUPLAND, G. & PERILLEUX, C. 2011. Cytokinin promotes flowering of Arabidopsis via transcriptional activation of the FT paralogue TSF. Plant J, 65, 972-9. 3. GUTIERREZ, L., MAURIAT, M., GUENIN, S., PELLOUX, J., LEFEBVRE, J. F., LOUVET, R., RUSTERUCCI, C., MORITZ, T., GUERINEAU, F., BELLINI, C. & VAN WUYTSWINKEL, O. 2008. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J., 6, 609-18. 4. HAN, S. & KIM, D. 2006. AtRTPrimer: database for Arabidopsis genome-wide homogeneous and specific RT-PCR primer-pairs. BMC bioinformatics, 7, 179. 5. LEE, H., YOO, S. J., LEE, J. H., KIM, W., YOO, S. K., FITZGERALD, H., CARRINGTON, J. C. & AHN, J. H. 2010. Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res., 38, 3081-93. 6. QU, W., SHEN, Z., ZHAO, D., YANG, Y. & ZHANG, C. 2009. MFEprimer: multiple factor evaluation of the specificity of PCR primers. Bioinformatics, 25, 276-8. 7. QU, W., ZHOU, Y., ZHANG, Y., LU, Y., WANG, X., ZHAO, D., YANG, Y. & ZHANG, C. 2012. MFEprimer-2.0: a fast thermodynamics-based program for checking PCR primer specificity. Nucleic Acids Res., 40, W205-8.