Population Genetics
advertisement

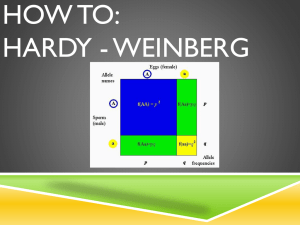
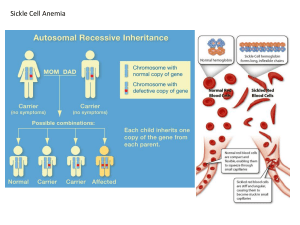
Population Genetics Allele Frequency – proportion of a specific allele relative to all other alleles in a population. Genotypic Frequency – proportion of a specific genotype relative to all other genotypes in a population. Phenotypic Frequency – proportion of a specific phenotype relative to all other phenotypes in a population. Calculating Allele Frequencies: Two alleles For two alleles A and a with relative proportions p and q, respectively, the sum of their proportions equals one: o p+q=1 o q=p–1 The frequency of an allele = frequency of homozygotes for that allele + ½ frequency of heterozygotes carrying that allele o If p = fA in a population, then p = fAA + ½(fAa) o If q = fa in a population, then q = faa + ½(fAa) Hardy-Weinberg Equilibrium –used to explain the persistence of recessive traits and the maintenance of deleterious genes (which should be selected against) in populations. Assumed conditions for this model: o Random mating o Infinitely large population o No mutation o No selection o No migration (in or out) Under these conditions, two alleles of an autosomal gene with allelic frequencies fA = p and fa = q will have genotypic frequencies of: o fAA = p2 o fAa = 2pq o faa = q2 These genotypic frequencies will persist (i.e. remain in equilibrium) as long as the five conditions of the model are met. Factors That Can Disturb H-W Equilibrium Non-random mating – e.g. assortive mating (like with like—sharing traits) or consanguineous mating. o Leads to increased homozygosity. Small Population Size – leads to fixation of genes (the allele in the population goes to 100%). All members of a population become homozygous for a particular allele. Also causes genetic drift. o Founder effect – one person of a small group with an uncommon allele moves to an isolated area; the allele becomes prominent. o Bottle-Neck effect – an abrupt decrease in population size (often due to an environmental cause) results in an increase in the prevalence (frequency) of alleles carried by the survivors. Mutation – introduction of new alleles to the population. Selection – due to reproductive or genetic fitness (refers to the tendency for alleles to increase in the population, often due to some advantage conferred). o Heterozygote advantage – causes mutations that result in disease (when homozygous) to increase in prevalence within a population, because those individuals who are heterozygous are given a survival advantage (e.g. sickle cell anemia and malaria). Gene Flow –flux of genes into or out of a population due to migration. Proof of H-W Equilibrium The model is based upon equivalence of random “mating” of genotypes (i.e. mating partners are selected irrespective of genotype) with random combination of gametes at fertilization. Therefore, if chance determines the genotypes of mating individuals, then the allele (A or a) carried by the egg or sperm is equally subject to chance: o (p + q)2 = (1)2 = p2 + 2pq + q2 Sperm Eggs p (A) q (a) p (A) p2 (AA) pq (Aa) q (a) pq (Aa) q2 (aa) Using H-W Equilibrium Given the frequency (q = 0.3) of a deleterious, recessive allele (i.e. allele a) in a population, and knowing that the population is in H-W equilibrium, it is possible to determine 1) the frequency of phenotypically normal individuals in the population, 2) the frequency of carriers of allele a in the population, and 3) the frequency of affected individuals in the population. 1. Frequency of Normal Phenotypes: a. Given that q = 0.3, we know p = 0.7 b. Amount of normal phenotypes = p2 + 2pq = (0.7)2 + 2(0.7)(0.3) = 0.91 2. Frequency of Carriers: a. Frequency of carriers = frequency of heterozygotes = 2pq b. 2pq = 2(0.7)(0.3) = 0.42 3. Frequency of Affected Individuals: a. Affected = aa genotype = q2 b. q2 = 0.09 Example 2 – given a disease frequency of 1/10,000 for a recessive, monogenic trait in a population that is in H-W equilibrium, what is the frequency of carriers in the population? q2 (faa) = 1/10,000 , therefore q = 1/100 = fa p = 1 – q = 99/100 f carrier = fAa = 2pq = 2 (0.99)(0.01) = ~0.02 Special Case – X-linked traits Males and females have different equilibrium frequencies due to gene dosage differences and sex chromosome differences. Because females carry two X chromosomes, allele frequencies of p + q = 1 result in genotype frequencies of p2 +2pq + q2 = 1 Males only have one X chromosome, so both allele and genotype frequencies are represented by p + q = 1 Problem: o Given a disease frequency of 1/10,000 for an X-linked recessive trait among males in a population in H-W equilibrium, 1) what is the expected frequency of carriers for the trait in this population? 2) what is the expected frequency of affected females? 1. Calculate the frequency of the two alleles (fXA = p and fXa = q). a. Most of the affected will be male, and because X chromosomes are inherited directly from the mother and Y chromosomes directly from the father, individuals will either be XAY (normal) found at frequency p, or XaY (affected) found at frequency q. b. The frequency of affected males is determined by the frequency of the Xa allele among females. c. q = 1/10,000 = fXa ~ frequency of affected males. d. p = 1 – q = 9,999/10,000 = fXA ~ frequency of unaffected males. e. Carriers of an X-linked recessive trait must be female, and in a population that is in H-W equilibrium, an X-linked trait behaves in females as if it were an autosomal recessive trait. f. Thus, frequency of carriers = frequency of heterozygous females = 2pq = 2 (9,999/10,000)(1/10,000) = ~0.0002 2. Because the frequency of affected females is <<<< frequency of affected males, p approaches 1.0 for rare, X-linked traits. Frequency of affected females = 0.00000001 (much < than observed frequency in males) and can be ignored. Mutation-Selection Equilibrium In populations, new alleles arise by mutation and are maintained or removed by selection. Fitness (f)– measure of the number of offspring of affected persons who survive to reproductive age, compared with an appropriate control group. o Fitness range is between 0-1. Coefficient of Selection (s) = 1 – f o Acts as a measure of loss of fitness. Ex: Achondroplastic dwarves will have ~20% of the number of offspring compared to those of normal stature. o Fitness = 0.2 and therefore the coefficient of selection = 0.8 Warfarin Resistance in Rodents – a model for how deleterious, recessive alleles can be maintained (Evolution in real time). Warfarin rodenticide introduced in the 1940s-50s. Mechanism was to block recycling of vitamin K (clotting cofactor), and rodents that ingested it would hemorrhage following injury. This model exhibits selection for a single-gene, vitamin K epoxide reductase complex subunit 1 (Vkorc1) – an allele that confers resistance to warfarin. When warfarin was used, it created selective pressure on rat populations for resistance to warfarin-induced bleeding. There was selection for the Vkorc1 resistance (R) allele, versus the susceptible allele (S). o Studies found relative fitness of: 0.37 for SS 1.00 for SR 0.68 for RR o Frequency of R allele did not approach 1, because RR homozygotes had a vitamin K deficiency. Fitness and Autosomal Dominant Disorders Autosomal dominant disorders are often associated with reduced fitness. In the case of autosomal dominant disorders, fitness is inversely associated with the proportion of patients with that disorder that have new mutant genes (as in a somatic mutation that was not inherited). In disorders with f = 0: o Patients with such disorders never reproduce, and all observed cases are caused by newly arising mutations. In disorders with f = 1: o Patients with such disorders have essentially normal reproductive fitness, and observed cases are most likely due to inheritance of a mutant allele, very rarely from newly arising mutations. Mutation-Selection Equilibrium and H-W Equilibrium Autosomal Dominant Traits: o For a mutant allele to remain in H-W equilibrium, the mutation rate / generation (μ) must be sufficient to balance the proportion of mutant alleles (q) lost in each generation from selection (s). For autosomal dominant traits: μ = sq = (1-f)q Autosomal Recessive Traits: o Selection against deleterious recessive mutations is much less influential on allele frequencies, because the frequency of affected individuals (faa) upon which selection acts represents a much smaller fraction of the population (This is the fallacy of eugenics). X-linked Recessive Traits: o If the phenotype is benign (i.e. affected males live to reproduce), then 1/3 of all mutant alleles will be in males (because they only have one X chromosome), while 2/3 of all mutant alleles will be in females. o Thus, the mutation rate / generation (μ) must equal the selection coefficient (s) multiplied by q/3 (because selection only acts against hemizygous males, not carrier females). μ = s(q/3) = (1-f) (q/3) o In the case of X-linked genetic lethality, where f~0 and s~1, each generation will lose 1/3 of all copies of the mutant allele. To maintain the observed disease incidence, mutant alleles lost by selection must be replaced by recurrent production of newly-arising mutations. Inbreeding Consanguineous mating: A mating between close relatives. Offspring from such a mating are considered inbred. Coefficient of Relationship – a measure of the proportion of genes shared by two related individuals. Often calculated for closely-related people who form a mating pair. In such a relationship, both individuals will have a certain proportion of alleles that are identical by descent (have a common history). Formula for the coefficient of relationship: o (1/2)n , where n = number of matings that connect related individuals. Example 1 – first cousins. o Number of mating pairs connecting = 3 (grandparents, parents of male cousin and parents of female cousin). (1/2)3 = 1/8 Example 2 – parent-child relationship o (1/2)1 = ½ The proportion of genes shared = the probability that a particular allele (such as a mutant allele causing a recessive trait) is shared. Coefficient of Inbreeding (F)– A measure of the probability that a homozygote has received both alleles from a common, recent ancestor. Also a measure of the proportion of loci for which a person is homozygous (i.e. both alleles are identical by descent). F = ½ coefficient of relationship o This formula represents P(inheriting a common allele) x P(other parent also passing on that allele) **This relationship only applies when there are low levels of inbreeding, and not to livestock breeding or inbred laboratory strains (requires more complex formula). Inbred Strains – produced by 20 or more generations of brother-sister matings. o After 20 generations of brother sister matings, 99% of loci will be homozygous. o Ex: albino and pigmented rats. Risk Assessment Risk Calculation Events in probability calculations are either: o mutually exclusive (“or” event, meaning addition of probabilities) o or independent (“and” event, meaning multiplication of probabilities). **Note: The sex of an offspring is an independent event: o P(male) ~ P(female) ~ 0.5 o Extremely important in calculating the risk of inheriting a deleterious phenotype. Bayesian Analysis: Used in genetics to assess risk. Calculation of risk requires a combination of several probability values. The prior probability distribution of an uncertain quantity “p” represents the probability of an uncertain event before any data is collected. o Ex: When constructing a pedigree for a trait that exhibits autosomal dominance, any affected individual has a prior probability of ½ for being a heterozygote (we are unsure if this person is nonheterozygous or heterozygous and affected by the dominant trait). o Provided that you have adequate family history, complete penetrance and can identify heterozygotes; it is a rule that in pedigrees exhibiting an autosomal dominant trait there is a 50% chance of passing on the mutant allele. Conditional probability is the probability that an event will occur, when another event is known to occur or to have occurred. In genetics, conditional probability is used to represent the probability that one event (that has occurred) accounts for another event. o Ex: If an individual is affected by an autosomal dominant trait, the prior probability of that individual being a heterozygote is ½. Conditionally, since we know that the individual is affected, he/she must be either a heterozygote or a homozygote for the mutant allele (each possibility with a probability of ½). Joint probability = prior probability x conditional probability. o Following the examples above, an affected individual has 0.5 chance of being a heterozygote, and conditionally there is a 0.5 chance of a heterozygous genotype being the CAUSE of the disease (the other 0.5 chance would be a homozygous genotype). o Therefore in this example, the joint probability = ½ * ½ = ¼ Posterior probability of a random event is the conditional probability that is assigned after relevant evidence is taken into account (i.e. evidence from a survey or study). o To calculate posterior probability, you must find the joint probabilities of two potential outcomes. The joint probability of each outcome is then divided by the sum of the joint probabilities from both outcomes: o Example – Pedigree For Huntington Disease (Autosomal Dominant) I-2 has Huntington Disease. It has been observed that 50% of heterozygotes manifest disease by age 50. In this example, we want to determine the probability that III-1 is heterozygous for Huntington Disease. To determine this, we must know the probability that II-1 carries the mutant allele. Since we do not know whether I-2 is heterozygous or homozygous, we assign a prior probability of ½ to the chance that she is heterozygous or nonheterozygous. In assessing the probability that II-1 is carrying the mutant allele, there is a conditional probability of 1 (certainty) that, if her mother is nonheterozygous, she is carrying the allele. o In other words, if her mother is not a heterozygote, she must be homozygous for the mutant allele (because she was affected), therefore it is certain that II-1 is a carrier. In assessing the probability that II-1 is carrying the mutant allele, there is a conditional probability of ½ that IF her mother (I-2) was a heterozygote, then II-2 is also a heterozygote (there is a ½ chance that she is homozygous recessive). The joint probabilities of these independent events is calculated by multiplying prior and conditional probabilities, and the posterior or relative possibilities are found by dividing each joint probability by the sum of both joint probabilities. Because II-1 has a P(1/3) for being heterozygous, and there is a P(1/2) of her passing on the mutant allele, the probability that III-1 is heterozygous is: o P = (1/3)(1/2) = 1/6